Calculating Global Coherence as a function of Glacial/Interglacial cycles#
This notebook lays out the details of how we use Pyleoclim to calculate global coherence between insolation and oxygen isotopes from a subset of records as a function of glacial/interglacial cycles.
The notebook is structured as follows:
Define a function that will be used to calculate correlations between records that contain large hiatuses
Define insolation curves using climlab
Define boundaries of interglacial/glacial periods use those defined in LR04
Break the Sanbao record into glacial/interglacial components and create two series object, one for each.
Analyze global coherence between insolation and the glacial/interglacial Sanbao series objects.
Repeat steps 4 and 5 for the oxygen isotope data from Buckeye Creek
Repeat steps 4 and 5 for oxygen isotope data from a marine sediment core in the Bay of Bengal
# Importing the necessary libraries
import pickle
import pyleoclim as pyleo
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
from climlab.solar.orbital import OrbitalTable
from climlab.solar.insolation import daily_insolation
from pylipd.lipd import LiPD
# Importing the data
with open("../../data/geo_ms_composite_dict.pkl", "rb") as handle:
geo_ms_composite_dict = pickle.load(handle)
with open("../../data/cmap_grouped.pkl", "rb") as handle:
cmap = pickle.load(handle)
with open("../../data/correlated_latitude.pkl", "rb") as handle:
correlated_latitude = pickle.load(handle)
Creating integrated summer insolation curve at 30N (picked arbitrarily):
# Creating insolation object
# array with specified kyears (can be plain numpy or xarray.DataArray)
years = np.arange(-1000, 1)
# subset of orbital parameters for specified time
orb = OrbitalTable.interp(kyear=years)
# Day numbers from June 1st to August 31st
jja_days = np.arange(152, 243)
lat = 30
days = jja_days
inso = daily_insolation(lat=lat, day=days, orb=orb).mean(dim="day")
inso_series = pyleo.Series(
time=0 - years[::-1],
value=inso[::-1],
time_name="Age",
time_unit="Kyr BP",
value_name=f"JJA Insolation {lat} N",
value_unit="W/m^2",
verbose=False,
)
Building MIS boundaries:
# Building the MIS boundaries
MIS_df = pd.read_table(
"https://lorraine-lisiecki.com/LR04_MISboundaries.txt",
skiprows=1,
header=0,
delim_whitespace=True,
nrows=25,
index_col="Boundary",
)
interglacial_to_glacial = [
"1/2",
"5/6",
"7/8",
"9/10",
"11/12",
"13/14",
"15/16",
"17/18",
"19/20",
]
glacial_to_interglacial = [
"4/5",
"6/7",
"8/9",
"10/11",
"12/13",
"14/15",
"16/17",
"18/19",
]
glacial_timing = [
(
MIS_df.loc[interglacial_to_glacial[idx]]["Age(ka)"],
MIS_df.loc[glacial_to_interglacial[idx]]["Age(ka)"],
)
for idx in range(len(glacial_to_interglacial))
]
interglacial_timing = [
(glacial_timing[idx - 1][1], glacial_timing[idx][0])
for idx in range(1, len(glacial_to_interglacial))
]
interglacial_timing.insert(0, (0, glacial_timing[0][0]))
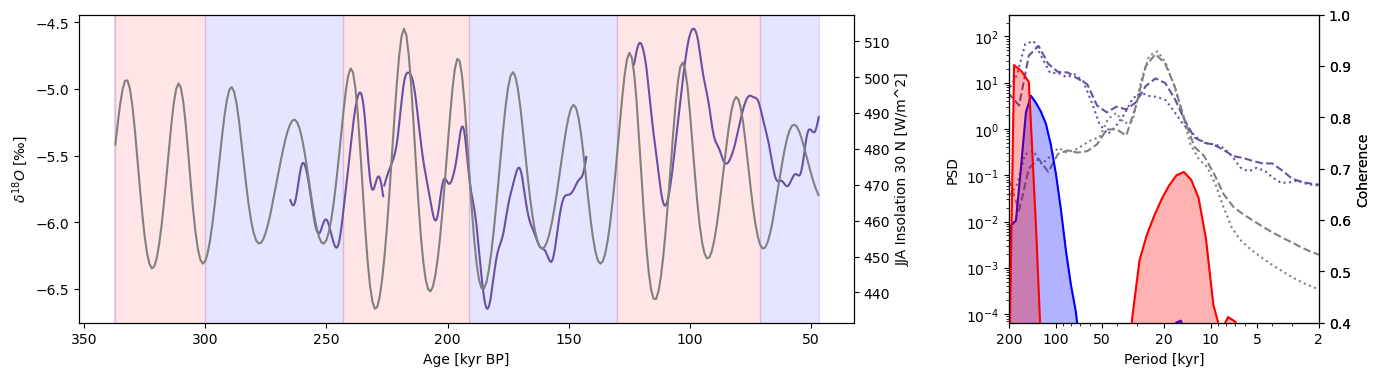
Sanbao Analysis#
Creating glacial/interglacial series objects using MIS boundaries:
# Creating interglacial series objects
series = geo_ms_composite_dict["Sanbao.China.2016"].convert_time_unit(
"kyrs BP"
) # .interp()
value = []
time = []
for interval in interglacial_timing:
series_interval = series.slice(interval)
if len(series_interval.time) > 1:
value.extend(series_interval.value)
time.extend(series_interval.time)
interglacial_series = series.copy()
interglacial_series.time = time
interglacial_series.value = value
# Creating glacial series objects
value = []
time = []
for interval in glacial_timing:
series_interval = series.slice(interval)
if len(series_interval.time) > 1:
value.extend(series_interval.value)
time.extend(series_interval.time)
glacial_series = series.copy()
glacial_series.time = time
glacial_series.value = value
Calculating global coherence and plotting:
# Calculating coherence and plotting the data
fig = plt.figure(figsize=(16, 4))
gs = fig.add_gridspec(nrows=1, ncols=3, wspace=0.5)
ax = fig.add_subplot(gs[0, 0:2])
ts = geo_ms_composite_dict["Sanbao.China.2016"]
ts.value_name = r"$\delta^{18}O$"
ts.value_unit = "‰"
ts_plot = ts.interp().filter(cutoff_scale=6)
max_age = max(ts.time)
min_age = min(ts.time)
glacial_intervals = []
interglacial_intervals = []
for glacial_end, glacial_start in glacial_timing:
if min_age < glacial_end < max_age or min_age < glacial_start < max_age:
glacial_intervals.append(
[max(min_age, glacial_end), min(max_age, glacial_start)]
)
for interglacial_end, interglacial_start in interglacial_timing:
if min_age < interglacial_end < max_age or min_age < interglacial_start < max_age:
interglacial_intervals.append(
[max(min_age, interglacial_end), min(max_age, interglacial_start)]
)
ts_plot.plot(ax=ax, color=cmap[ts.label], legend=False)
ax.invert_xaxis()
for glacial_interval in glacial_intervals:
ax.axvspan(glacial_interval[0], glacial_interval[1], color="b", alpha=0.1)
for interglacial_interval in interglacial_intervals:
ax.axvspan(interglacial_interval[0], interglacial_interval[1], color="r", alpha=0.1)
ax_twin = ax.twinx()
ax_twin.grid(False)
inso_series.slice((min(ts.time), max(ts.time))).plot(ax=ax_twin, color="grey")
ax2 = fig.add_subplot(gs[0, 2])
glacial_coh_real = glacial_series.global_coherence(inso_series)
glacial_coh_real.plot(
ax=ax2,
spec1_plot_kwargs={"color": cmap[ts.label], "linestyle": "dotted"},
spec2_plot_kwargs={"color": "grey", "linestyle": "dotted"},
coh_line_color="blue",
fill_color="blue",
legend=False,
)
interglacial_coh_real = interglacial_series.global_coherence(inso_series)
interglacial_coh_real.plot(
ax=ax2,
spec1_plot_kwargs={"color": cmap[ts.label], "linestyle": "dashed"},
spec2_plot_kwargs={"color": "grey", "linestyle": "dashed"},
coh_line_color="red",
fill_color="red",
legend=False,
)
<Axes: xlabel='Period [kyr]', ylabel='PSD'>
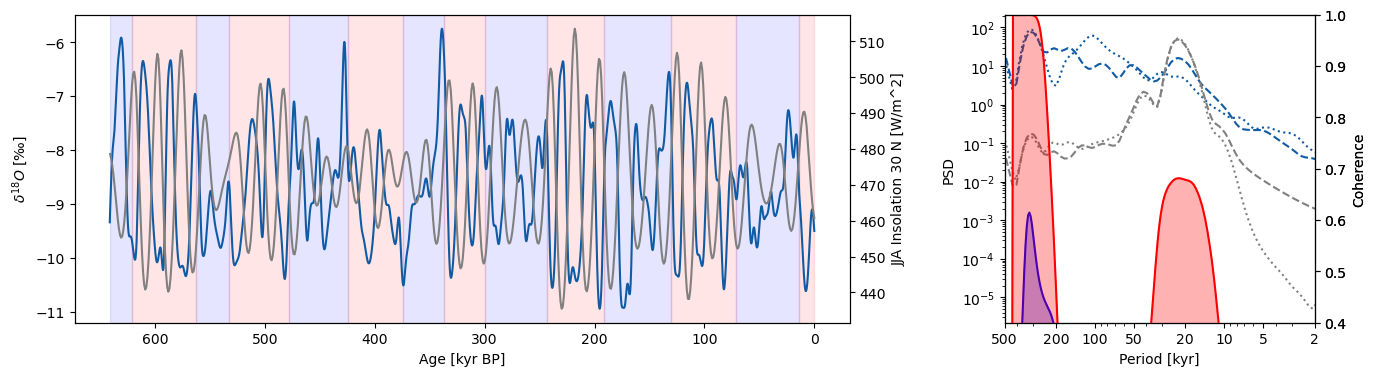
Buckeye Creek#
# Loading smoothed plotting dict, necessary here
with open("../../data/smooth_plotting_series_dict.pkl", "rb") as handle:
smoothed_plotting_series = pickle.load(handle)
Creating glacial/interglacial series for Buckeye Creek oxygen isotope records:
# Creating the interglacial series
series = geo_ms_composite_dict["BuckeyeCreek.WestVirginia.2019"].convert_time_unit(
"kyrs BP"
) # .interp()
value = []
time = []
for interval in interglacial_timing:
series_interval = series.slice(interval)
if len(series_interval.time) > 1:
value.extend(series_interval.value)
time.extend(series_interval.time)
interglacial_series = series.copy()
interglacial_series.time = time
interglacial_series.value = value
# Creating the glacial series
value = []
time = []
for interval in glacial_timing:
series_interval = series.slice(interval)
if len(series_interval.time) > 1:
value.extend(series_interval.value)
time.extend(series_interval.time)
glacial_series = series.copy()
glacial_series.time = time
glacial_series.value = value
Calculating global coherence and plotting:
# Calculating coherence and plotting the data
fig = plt.figure(figsize=(16, 4))
gs = fig.add_gridspec(nrows=1, ncols=3, wspace=0.5)
ax = fig.add_subplot(gs[0, 0:2])
ts = geo_ms_composite_dict["BuckeyeCreek.WestVirginia.2019"].convert_time_unit(
"kyrs BP"
)
ts_plot = smoothed_plotting_series["BuckeyeCreek.WestVirginia.2019"].convert_time_unit(
"kyrs BP"
)
ts.value_name = r"$\delta^{18}O$"
ts.value_unit = "‰"
max_age = max(ts.time)
min_age = min(ts.time)
glacial_intervals = []
interglacial_intervals = []
for glacial_end, glacial_start in glacial_timing:
if min_age < glacial_end < max_age or min_age < glacial_start < max_age:
glacial_intervals.append(
[max(min_age, glacial_end), min(max_age, glacial_start)]
)
for interglacial_end, interglacial_start in interglacial_timing:
if min_age < interglacial_end < max_age or min_age < interglacial_start < max_age:
interglacial_intervals.append(
[max(min_age, interglacial_end), min(max_age, interglacial_start)]
)
ts_plot.plot(ax=ax, color=cmap[ts.label], legend=False)
ax.invert_xaxis()
ax.set_xlabel("Age [kyr BP]")
for glacial_interval in glacial_intervals:
ax.axvspan(glacial_interval[0], glacial_interval[1], color="b", alpha=0.1)
for interglacial_interval in interglacial_intervals:
ax.axvspan(interglacial_interval[0], interglacial_interval[1], color="r", alpha=0.1)
ax_twin = ax.twinx()
ax_twin.grid(False)
inso_series.slice((min(ts.time), max(ts.time))).plot(ax=ax_twin, color="grey")
ax2 = fig.add_subplot(gs[0, 2])
glacial_coh_real = glacial_series.global_coherence(inso_series)
glacial_coh_real.plot(
ax=ax2,
spec1_plot_kwargs={"color": cmap[ts.label], "linestyle": "dotted"},
spec2_plot_kwargs={"color": "grey", "linestyle": "dotted"},
coh_line_color="blue",
fill_color="blue",
legend=False,
)
interglacial_coh_real = interglacial_series.global_coherence(inso_series)
interglacial_coh_real.plot(
ax=ax2,
spec1_plot_kwargs={"color": cmap[ts.label], "linestyle": "dashed"},
spec2_plot_kwargs={"color": "grey", "linestyle": "dashed"},
coh_line_color="red",
fill_color="red",
legend=False,
)
<Axes: xlabel='Period [kyr]', ylabel='PSD'>
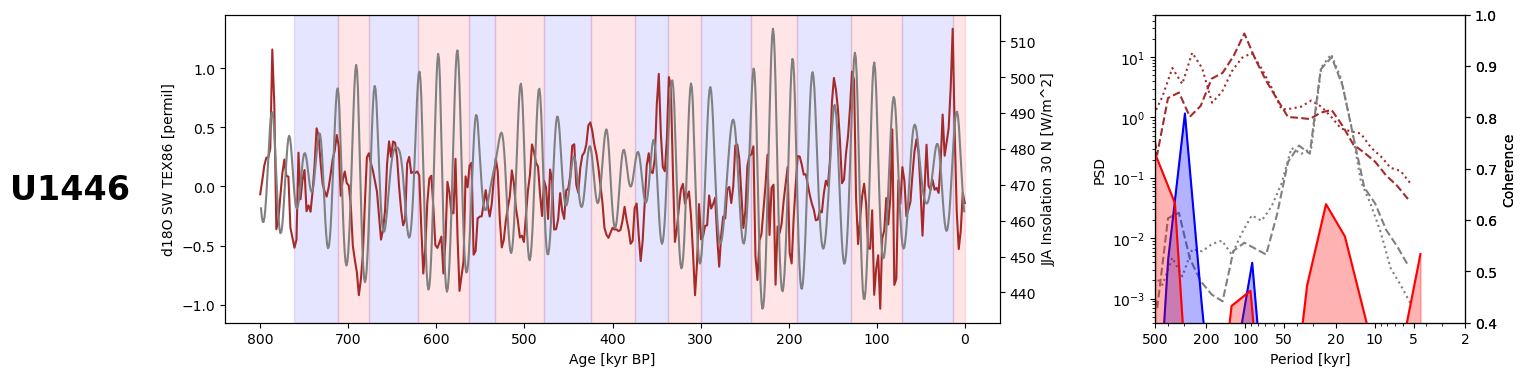
U1446#
Loading U1446 from LiPD, as we haven’t used this record before:
# Loading the U1446 marine sediment data
lipd_path = "../../data/marine_sediments/U1446.IndianOcean.2021.lpd"
L = LiPD()
if __name__ == "__main__":
L.load(lipd_path)
Loading 1 LiPD files
0%| | 0/1 [00:00<?, ?it/s]
100%|████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 1/1 [00:00<00:00, 35.80it/s]
Loaded..
Creating a series object from the reconstructed oxygen isotope composition of seawater (we use the TEX86 reconstruction, see the original publication for details):
# Loading the U1446 marine sediment data
ms_dict = {}
for _, row in L.get_timeseries_essentials().iterrows():
time = row["time_values"]
time_name = row["time_variableName"]
time_unit = row["time_units"]
value = row["paleoData_values"]
value_name = row["paleoData_variableName"]
value_unit = row["paleoData_units"]
lat = row["geo_meanLat"]
lon = row["geo_meanLon"]
if value_name == "d18O SW TEX86":
series = pyleo.GeoSeries(
time=time,
value=value,
time_name=time_name,
time_unit=time_unit,
value_name=value_name,
value_unit=value_unit,
lat=lat,
lon=lon,
archiveType="Marine Sediment",
verbose=False,
).interp()
sw_series = series.slice([0, 800])
break
Creating glacial and interglacial series from the U1446 series:
# Creating the interglacial series
series = sw_series
value = []
time = []
for interval in interglacial_timing:
series_interval = series.slice(interval)
if len(series_interval.time) > 1:
value.extend(series_interval.value)
time.extend(series_interval.time)
interglacial_series = series.copy()
interglacial_series.time = time
interglacial_series.value = value
# Creating the glacial series
value = []
time = []
for interval in glacial_timing:
series_interval = series.slice(interval)
if len(series_interval.time) > 1:
value.extend(series_interval.value)
time.extend(series_interval.time)
glacial_series = series.copy()
glacial_series.time = time
glacial_series.value = value
Calculating global coherence and plotting:
# Calculating coherence and plotting the data
fig = plt.figure(figsize=(16, 4))
gs = fig.add_gridspec(nrows=1, ncols=3, wspace=0.5)
ax = fig.add_subplot(gs[0, 0:2])
ts = series
ts_plot = series.filter(cutoff_scale=5)
ts.value_name = r"$\delta^{18}O$"
ts.value_unit = "‰"
max_age = max(ts.time)
min_age = min(ts.time)
glacial_intervals = []
interglacial_intervals = []
for glacial_end, glacial_start in glacial_timing:
if min_age < glacial_end < max_age or min_age < glacial_start < max_age:
glacial_intervals.append(
[max(min_age, glacial_end), min(max_age, glacial_start)]
)
for interglacial_end, interglacial_start in interglacial_timing:
if min_age < interglacial_end < max_age or min_age < interglacial_start < max_age:
interglacial_intervals.append(
[max(min_age, interglacial_end), min(max_age, interglacial_start)]
)
ts_plot.plot(ax=ax, color="brown", legend=False)
ax.invert_xaxis()
for glacial_interval in glacial_intervals:
ax.axvspan(glacial_interval[0], glacial_interval[1], color="b", alpha=0.1)
for interglacial_interval in interglacial_intervals:
ax.axvspan(interglacial_interval[0], interglacial_interval[1], color="r", alpha=0.1)
ax_twin = ax.twinx()
ax_twin.grid(False)
inso_series.slice((min(ts.time), max(ts.time))).plot(ax=ax_twin, color="grey")
ax2 = fig.add_subplot(gs[0, 2])
glacial_coh_real = glacial_series.global_coherence(inso_series)
glacial_coh_real.plot(
ax=ax2,
spec1_plot_kwargs={"color": "brown", "linestyle": "dotted"},
spec2_plot_kwargs={"color": "grey", "linestyle": "dotted"},
coh_line_color="blue",
fill_color="blue",
legend=False,
)
interglacial_coh_real = interglacial_series.global_coherence(inso_series)
interglacial_coh_real.plot(
ax=ax2,
spec1_plot_kwargs={"color": "brown", "linestyle": "dashed"},
spec2_plot_kwargs={"color": "grey", "linestyle": "dashed"},
coh_line_color="red",
fill_color="red",
legend=False,
)
ax.text(
-0.2,
0.4,
"U1446",
transform=ax.transAxes,
ha="center",
fontsize=24,
fontweight="bold",
)
ax2.set_xlim(500, 0)
ax2.set_xticks(
[500, 200, 100, 50, 20, 10, 5, 2],
)
[<matplotlib.axis.XTick at 0x339729790>,
<matplotlib.axis.XTick at 0x33e0cc410>,
<matplotlib.axis.XTick at 0x3397e0590>,
<matplotlib.axis.XTick at 0x3397e1a50>,
<matplotlib.axis.XTick at 0x3397e3890>,
<matplotlib.axis.XTick at 0x3397e5e90>,
<matplotlib.axis.XTick at 0x339773dd0>,
<matplotlib.axis.XTick at 0x339a55810>]