Assembling the proxy database from the LiPDVerse#
⚠️ Warning
This notebook contains a cell that starts a widget to help with identifying duplicates in the database. We strongly advise that you do not run the widget yourself unless you intend to go through the cleaning process. A list of removed datasets (TSiDS) and the reason they were removed or kept is stored in csv files in this repository. The resulting cleaned database has also been saved in a pickle file.
Goal#
The goal of this notebook is to showcase data querying on the LiPDverser to create the proxy database
Data Description#
This notebook makes use of the LiPDVerse database, which contains datasets from various working groups. For a list of these compilations, see this page.
In particular, we will be using PAGES2kTemperature, iso-2k, and CoralHydro2k compilations for this demonstration.
PAGES2k: PAGES2k Consortium., Emile-Geay, J., McKay, N. et al. A global multiproxy database for temperature reconstructions of the Common Era. Sci Data 4, 170088 (2017). doi:10.1038/sdata.2017.88
iso2k: Konecky, B. L., McKay, N. P., Churakova (Sidorova), O. V., Comas-Bru, L., Dassié, E. P., DeLong, K. L., Falster, G. M., Fischer, M. J., Jones, M. D., Jonkers, L., Kaufman, D. S., Leduc, G., Managave, S. R., Martrat, B., Opel, T., Orsi, A. J., Partin, J. W., Sayani, H. R., Thomas, E. K., Thompson, D. M., Tyler, J. J., Abram, N. J., Atwood, A. R., Cartapanis, O., Conroy, J. L., Curran, M. A., Dee, S. G., Deininger, M., Divine, D. V., Kern, Z., Porter, T. J., Stevenson, S. L., von Gunten, L., and Iso2k Project Members: The Iso2k database: a global compilation of paleo-δ18O and δ2H records to aid understanding of Common Era climate, Earth Syst. Sci. Data, 12, 2261–2288, https://doi.org/10.5194/essd-12-2261-2020, 2020.
CoralHydro2k: Walter, R. M., H. R. Sayani, T. Felis, K. M. Cobb, N. J. Abram, A. K. Arzey, A. R. Atwood, L. D. Brenner, E. P. Dassi ́e, K. L. DeLong, B. Ellis, J. Emile-Geay, M. J. Fischer, N. F. Goodkin, J. A. Hargreaves, K. H. Kilbourne, H. Krawczyk, N. P. McKay, A. L. Moore, S. A. Murty, M. R. Ong, R. D. Ramos, E. V. Reed, D. Samanta, S. C. Sanchez, J. Zinke, and the PAGES CoralHydro2k Project Members (2023), The CoralHydro2k database: a global, actively curated compilation of coral δ18O and Sr / Ca proxy records of tropical ocean hydrology and temperature for the Common Era, Earth System Science Data, 15(5), 2081–2116, doi:10.5194/essd-15-2081-2023.
Let’s start by downloading the datasets.
# URL Links to download the data
pages2k_url = "https://lipdverse.org/Pages2kTemperature/current_version/Pages2kTemperature2_2_0.zip"
iso2k_url = "https://lipdverse.org/iso2k/current_version/iso2k1_1_2.zip"
coral2k_url ="https://lipdverse.org/CoralHydro2k/current_version/CoralHydro2k1_0_1.zip"
urls = [pages2k_url,iso2k_url,coral2k_url]
folders = ['Pages2k', 'iso2k', 'coral2k']
# import the necessary packages
import os
import requests
import zipfile
from pathlib import Path
## PAGES2k
# Define folder path and URL
for idx,url in enumerate(urls):
folder_path = Path(f"./data/{folders[idx]}")
zip_path = folder_path.with_suffix('.zip')
# Step 1: Check if folder exists, create if not
folder_path.mkdir(parents=True, exist_ok=True)
# Step 2: Check if folder is empty
if not any(folder_path.iterdir()):
print("Folder is empty. Downloading data...")
# Step 3: Download zip file
response = requests.get(url)
with open(zip_path, 'wb') as f:
f.write(response.content)
print(f"Downloaded ZIP to {zip_path}")
# Step 4: Extract zip contents into folder
with zipfile.ZipFile(zip_path, 'r') as zip_ref:
zip_ref.extractall(folder_path)
print(f"Extracted contents to {folder_path}")
Folder is empty. Downloading data...
Downloaded ZIP to data/Pages2k.zip
Extracted contents to data/Pages2k
Folder is empty. Downloading data...
Downloaded ZIP to data/iso2k.zip
Extracted contents to data/iso2k
Folder is empty. Downloading data...
Downloaded ZIP to data/coral2k.zip
Extracted contents to data/coral2k
Exploring the records in each of the compilations#
Let’s start with importing the needed packages:
import os
# General data manipulation libraries
import numpy as np
import pandas as pd
# To manipulate LiPD files
from pylipd.lipd import LiPD
import json
# Scientific toolboxes
import pyleoclim as pyleo
import seaborn as sns
import cfr
from scipy import stats
#To be able to compare timeseries
from scipy.stats import zscore
from dtaidistance import dtw
from scipy.stats import pearsonr, zscore
import matplotlib.pyplot as plt
import itertools
#For saving
import pickle
PAGES2k#
Let’s start with the PAGES2k dataset. We can load the entire folder into a pylipd.LiPD object:
D_pages2k = LiPD()
D_pages2k.load_from_dir('./data/Pages2k/')
Loading 647 LiPD files
100%|██████████| 647/647 [00:11<00:00, 54.47it/s]
Loaded..
Let’s create a query to gather all the necessary information. Since PyLiPD is build upon the rdflib library and LiPD is defined as a child of the Graph class in rdflib, it is possible to directly query a LiPD object using a SPARQL query as in this tutorial:
query = """ PREFIX le: <http://linked.earth/ontology#>
PREFIX wgs84: <http://www.w3.org/2003/01/geo/wgs84_pos#>
PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>
SELECT ?dataSetName ?compilationName ?archiveType ?geo_meanLat ?geo_meanLon ?geo_meanElev
?paleoData_variableName ?paleoData_standardName ?paleoData_values ?paleoData_units
?paleoData_proxy ?paleoData_proxyGeneral ?paleoData_seasonality ?paleoData_interpName ?paleoData_interpRank
?TSID ?time_variableName ?time_standardName ?time_values
?time_units where{
?ds a le:Dataset .
?ds le:hasName ?dataSetName .
# Get the archive
OPTIONAL{
?ds le:hasArchiveType ?archiveTypeObj .
?archiveTypeObj rdfs:label ?archiveType .
}
# Geographical information
?ds le:hasLocation ?loc .
OPTIONAL{?loc wgs84:lat ?geo_meanLat .}
OPTIONAL{?loc wgs84:long ?geo_meanLon .}
OPTIONAL{?loc wgs84:alt ?geo_meanElev .}
# PaleoData
?ds le:hasPaleoData ?data .
?data le:hasMeasurementTable ?table .
?table le:hasVariable ?var .
# Name
?var le:hasName ?paleoData_variableName .
?var le:hasStandardVariable ?variable_obj .
?variable_obj rdfs:label ?paleoData_standardName .
#Values
?var le:hasValues ?paleoData_values .
#Seasonality
OPTIONAL{?var le:seasonality ?paleoData_seasonality} .
#Units
OPTIONAL{
?var le:hasUnits ?paleoData_unitsObj .
?paleoData_unitsObj rdfs:label ?paleoData_units .
}
#Proxy information
OPTIONAL{
?var le:hasProxy ?paleoData_proxyObj .
?paleoData_proxyObj rdfs:label ?paleoData_proxy .
}
OPTIONAL{
?var le:hasProxyGeneral ?paleoData_proxyGeneralObj .
?paleoData_proxyGeneralObj rdfs:label ?paleoData_proxyGeneral .
}
# Compilation
?var le:partOfCompilation ?compilation .
?compilation le:hasName ?compilationName .
FILTER (?compilationName = "Pages2kTemperature").
?var le:useInGlobalTemperatureAnalysis True .
# TSiD (should all have them)
OPTIONAL{
?var le:hasVariableId ?TSID
} .
# Interpretation (might be an optional field)
OPTIONAL{
?var le:hasInterpretation ?interp .
?interp le:hasVariable ?interpvar .
BIND(REPLACE(STR(?interpvar), "http://linked.earth/ontology/interpretation#", "") AS ?interpretedVariable_Fallback)
OPTIONAL { ?interpvar rdfs:label ?interpretedVariable_Label } # Use a temporary variable for the label
BIND(COALESCE(?interpretedVariable_Label, ?interpretedVariable_Fallback) AS ?paleoData_interpName) # COALESCE into the final variable
}
OPTIONAL{
?var le:hasInterpretation ?interp .
?interp le:hasRank ?paleoData_interpRank .}
OPTIONAL{
?var le:hasInterpretation ?interp .
?interp le:hasSeasonality ?seasonalityURI .
BIND(REPLACE(STR(?seasonalityURI), "http://linked.earth/ontology/interpretation#", "") AS ?seasonality_Fallback)
OPTIONAL { ?seasonalityURI rdfs:label ?seasonalityLabel }
BIND(COALESCE(?seasonalityLabel, ?seasonality_Fallback) AS ?paleoData_seasonality)
}
#Time information
?table le:hasVariable ?timevar .
?timevar le:hasName ?time_variableName .
?timevar le:hasStandardVariable ?timevar_obj .
?timevar_obj rdfs:label ?time_standardName .
VALUES ?time_standardName {"year"} .
?timevar le:hasValues ?time_values .
OPTIONAL{
?timevar le:hasUnits ?time_unitsObj .
?time_unitsObj rdfs:label ?time_units .
}
}"""
qres, df_pages2k = D_pages2k.query(query)
df_pages2k.head()
| dataSetName | compilationName | archiveType | geo_meanLat | geo_meanLon | geo_meanElev | paleoData_variableName | paleoData_standardName | paleoData_values | paleoData_units | paleoData_proxy | paleoData_proxyGeneral | paleoData_seasonality | paleoData_interpName | paleoData_interpRank | TSID | time_variableName | time_standardName | time_values | time_units | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Ant-WDC05A.Steig.2013 | Pages2kTemperature | Glacier ice | -79.46 | -112.09 | 1806.0 | d18O | d18O | [-33.32873325, -35.6732, -33.1574, -34.2854, -... | permil | d18O | None | Annual | temperature | NaN | Ant_030 | year | year | [2005, 2004, 2003, 2002, 2001, 2000, 1999, 199... | yr AD |
| 1 | NAm-MtLemon.Briffa.2002 | Pages2kTemperature | Wood | 32.50 | -110.80 | 2700.0 | MXD | MXD | [0.968, 0.962, 1.013, 0.95, 1.008, 0.952, 1.02... | None | maximum latewood density | dendrophysical | Summer | temperature | NaN | NAm_568 | year | year | [1568, 1569, 1570, 1571, 1572, 1573, 1574, 157... | yr AD |
| 2 | Arc-Arjeplog.Bjorklund.2014 | Pages2kTemperature | Wood | 66.30 | 18.20 | 800.0 | density | density | [-0.829089212152348, -0.733882889924006, -0.89... | None | maximum latewood density | dendrophysical | Jun-Aug | temperature | NaN | Arc_060 | year | year | [1200, 1201, 1202, 1203, 1204, 1205, 1206, 120... | yr AD |
| 3 | Asi-CHIN019.Li.2010 | Pages2kTemperature | Wood | 29.15 | 99.93 | 2150.0 | ringWidth | ringWidth | [1.465, 1.327, 1.202, 0.757, 1.094, 1.006, 1.2... | None | ring width | dendrophysical | Annual | temperature | NaN | Asia_041 | year | year | [1509, 1510, 1511, 1512, 1513, 1514, 1515, 151... | yr AD |
| 4 | NAm-Landslide.Luckman.2006 | Pages2kTemperature | Wood | 60.20 | -138.50 | 800.0 | ringWidth | ringWidth | [1.123, 0.841, 0.863, 1.209, 1.139, 1.056, 0.8... | None | ring width | dendrophysical | Summer | temperature | NaN | NAm_1876 | year | year | [913, 914, 915, 916, 917, 918, 919, 920, 921, ... | yr AD |
Let’s have a look at the resulting data.
df_pages2k.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 691 entries, 0 to 690
Data columns (total 20 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 dataSetName 691 non-null object
1 compilationName 691 non-null object
2 archiveType 691 non-null object
3 geo_meanLat 691 non-null float64
4 geo_meanLon 691 non-null float64
5 geo_meanElev 684 non-null float64
6 paleoData_variableName 691 non-null object
7 paleoData_standardName 691 non-null object
8 paleoData_values 691 non-null object
9 paleoData_units 283 non-null object
10 paleoData_proxy 691 non-null object
11 paleoData_proxyGeneral 500 non-null object
12 paleoData_seasonality 691 non-null object
13 paleoData_interpName 691 non-null object
14 paleoData_interpRank 1 non-null float64
15 TSID 691 non-null object
16 time_variableName 691 non-null object
17 time_standardName 691 non-null object
18 time_values 691 non-null object
19 time_units 691 non-null object
dtypes: float64(4), object(16)
memory usage: 108.1+ KB
df_pages2k['paleoData_interpName'].unique()
array(['temperature'], dtype=object)
We have 691 timeseries that represent temperature, consistent with the information from the LiPDverse.
iso2k#
Let’s do something similar for the iso2k database:
D_iso2k = LiPD()
D_iso2k.load_from_dir('./data/iso2k')
Loading 509 LiPD files
100%|██████████| 509/509 [00:14<00:00, 34.94it/s]
Loaded..
query_iso = """ PREFIX le: <http://linked.earth/ontology#>
PREFIX wgs84: <http://www.w3.org/2003/01/geo/wgs84_pos#>
PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>
SELECT ?dataSetName ?compilationName ?archiveType ?geo_meanLat ?geo_meanLon ?geo_meanElev
?paleoData_variableName ?paleoData_standardName ?paleoData_values ?paleoData_units
?paleoData_proxy ?paleoData_proxyGeneral ?paleoData_seasonality ?paleoData_interpName ?paleoData_interpRank
?TSID ?time_variableName ?time_standardName ?time_values
?time_units where{
?ds a le:Dataset .
?ds le:hasName ?dataSetName .
# Get the archive
OPTIONAL{
?ds le:hasArchiveType ?archiveTypeObj .
?archiveTypeObj rdfs:label ?archiveType .
}
# Geographical information
?ds le:hasLocation ?loc .
OPTIONAL{?loc wgs84:lat ?geo_meanLat .}
OPTIONAL{?loc wgs84:long ?geo_meanLon .}
OPTIONAL{?loc wgs84:alt ?geo_meanElev .}
# PaleoData
?ds le:hasPaleoData ?data .
?data le:hasMeasurementTable ?table .
?table le:hasVariable ?var .
# Name
?var le:hasName ?paleoData_variableName .
?var le:hasStandardVariable ?variable_obj .
?variable_obj rdfs:label ?paleoData_standardName .
#Values
?var le:hasValues ?paleoData_values .
#Seasonality
OPTIONAL{?var le:seasonality ?paleoData_seasonality} .
#Units
OPTIONAL{
?var le:hasUnits ?paleoData_unitsObj .
?paleoData_unitsObj rdfs:label ?paleoData_units .
}
#Proxy information
OPTIONAL{
?var le:hasProxy ?paleoData_proxyObj .
?paleoData_proxyObj rdfs:label ?paleoData_proxy .
}
OPTIONAL{
?var le:hasProxyGeneral ?paleoData_proxyGeneralObj .
?paleoData_proxyGeneralObj rdfs:label ?paleoData_proxyGeneral .
}
# Compilation
?var le:partOfCompilation ?compilation .
?compilation le:hasName ?compilationName .
FILTER (?compilationName = "iso2k").
?var le:useInGlobalTemperatureAnalysis True .
# TSiD (should all have them)
OPTIONAL{
?var le:hasVariableId ?TSID
} .
# Interpretation (might be an optional field)
OPTIONAL{
?var le:hasInterpretation ?interp .
?interp le:hasVariable ?interpvar .
BIND(REPLACE(STR(?interpvar), "http://linked.earth/ontology/interpretation#", "") AS ?interpretedVariable_Fallback)
OPTIONAL { ?interpvar rdfs:label ?interpretedVariable_Label } # Use a temporary variable for the label
BIND(COALESCE(?interpretedVariable_Label, ?interpretedVariable_Fallback) AS ?paleoData_interpName) # COALESCE into the final variable
}
OPTIONAL{
?var le:hasInterpretation ?interp .
?interp le:hasRank ?paleoData_interpRank .}
OPTIONAL{
?var le:hasInterpretation ?interp .
?interp le:hasSeasonality ?seasonalityURI .
BIND(REPLACE(STR(?seasonalityURI), "http://linked.earth/ontology/interpretation#", "") AS ?seasonality_Fallback)
OPTIONAL { ?seasonalityURI rdfs:label ?seasonalityLabel }
BIND(COALESCE(?seasonalityLabel, ?seasonality_Fallback) AS ?paleoData_seasonality)
}
#Time information
?table le:hasVariable ?timevar .
?timevar le:hasName ?time_variableName .
?timevar le:hasStandardVariable ?timevar_obj .
?timevar_obj rdfs:label ?time_standardName .
VALUES ?time_standardName {"year"} .
?timevar le:hasValues ?time_values .
OPTIONAL{
?timevar le:hasUnits ?time_unitsObj .
?time_unitsObj rdfs:label ?time_units .
}
}"""
qres, df_iso2k = D_iso2k.query(query_iso)
df_iso2k.head()
| dataSetName | compilationName | archiveType | geo_meanLat | geo_meanLon | geo_meanElev | paleoData_variableName | paleoData_standardName | paleoData_values | paleoData_units | paleoData_proxy | paleoData_proxyGeneral | paleoData_seasonality | paleoData_interpName | paleoData_interpRank | TSID | time_variableName | time_standardName | time_values | time_units | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | LS16STCL | iso2k | Lake sediment | 50.830 | -116.39 | 1126.0 | d18O | d18O | [-7.81, -5.91, -9.03, -5.35, -5.61, -5.98, -5.... | permil | d18O | None | Winter | effectivePrecipitation | NaN | LPD7dc5b9ba-dup-dup | year | year | [2009.0, 2008.3, 2007.8, 2007.4, 2007.0, 2006.... | yr AD |
| 1 | LS16STCL | iso2k | Lake sediment | 50.830 | -116.39 | 1126.0 | d18O | d18O | [-7.81, -5.91, -9.03, -5.35, -5.61, -5.98, -5.... | permil | d18O | None | None | effectivePrecipitation | 3.0 | LPD7dc5b9ba-dup-dup | year | year | [2009.0, 2008.3, 2007.8, 2007.4, 2007.0, 2006.... | yr AD |
| 2 | CO00URMA | iso2k | Coral | 0.933 | 173.00 | 6.0 | d18O | d18O | [-4.8011, -4.725, -4.6994, -4.86, -5.0886, -5.... | permil | d18O | None | subannual | temperature | 1.0 | Ocean2kHR_177_iso2k | year | year | [1994.5, 1994.33, 1994.17, 1994.0, 1993.83, 19... | yr AD |
| 3 | CO00URMA | iso2k | Coral | 0.933 | 173.00 | 6.0 | d18O | d18O | [-4.8011, -4.725, -4.6994, -4.86, -5.0886, -5.... | permil | d18O | None | subannual | temperature | 1.0 | Ocean2kHR_177_iso2k | year | year | [1994.5, 1994.33, 1994.17, 1994.0, 1993.83, 19... | yr AD |
| 4 | CO00URMA | iso2k | Coral | 0.933 | 173.00 | 6.0 | d18O | d18O | [-4.8011, -4.725, -4.6994, -4.86, -5.0886, -5.... | permil | d18O | None | subannual | temperature | 1.0 | Ocean2kHR_177_iso2k | year | year | [1994.5, 1994.33, 1994.17, 1994.0, 1993.83, 19... | yr AD |
df_iso2k['paleoData_interpName'].unique()
array(['effectivePrecipitation', 'temperature', 'seawaterIsotope',
'precipitationIsotope', 'hydrologicBalance', 'salinity'],
dtype=object)
Despite using the useinGlobalTemperatureAnalysis flag, there seems to be more than 1 interpretation for several of the variables. Let’s first filer for interpretation associated with temperature and remove duplicate entries based on TSiD:
df_iso2k_filt = df_iso2k[df_iso2k['paleoData_interpName']=='temperature'].drop_duplicates(subset='TSID', keep='first')
df_iso2k_filt.head()
| dataSetName | compilationName | archiveType | geo_meanLat | geo_meanLon | geo_meanElev | paleoData_variableName | paleoData_standardName | paleoData_values | paleoData_units | paleoData_proxy | paleoData_proxyGeneral | paleoData_seasonality | paleoData_interpName | paleoData_interpRank | TSID | time_variableName | time_standardName | time_values | time_units | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | CO00URMA | iso2k | Coral | 0.9330 | 173.0000 | 6.0 | d18O | d18O | [-4.8011, -4.725, -4.6994, -4.86, -5.0886, -5.... | permil | d18O | None | subannual | temperature | 1.0 | Ocean2kHR_177_iso2k | year | year | [1994.5, 1994.33, 1994.17, 1994.0, 1993.83, 19... | yr AD |
| 11 | CO05KUBE | iso2k | Coral | 32.4670 | -64.7000 | -12.0 | d18O | d18O | [-4.15, -3.66, -3.69, -4.07, -3.95, -4.12, -3.... | permil | d18O | None | subannual | temperature | 1.0 | Ocean2kHR_105_iso2k | year | year | [1983.21, 1983.13, 1983.04, 1982.96, 1982.88, ... | yr AD |
| 13 | IC13THQU | iso2k | Glacier ice | -13.9333 | -70.8333 | 5670.0 | d18O | d18O | [-18.5905, -16.3244, -16.2324, -17.0112, -18.6... | permil | d18O | None | Winter | temperature | NaN | SAm_035_iso2k | year | year | [2009, 2008, 2007, 2006, 2005, 2004, 2003, 200... | yr AD |
| 15 | CO01TUNG | iso2k | Coral | -5.2170 | 145.8170 | -3.0 | d18O | d18O | [-4.875, -4.981, -5.166, -5.06, -4.942, -4.919... | permil | d18O | None | subannual | temperature | NaN | Ocean2kHR_141_iso2k | year | year | [1993.042, 1992.792, 1992.542, 1992.292, 1992.... | yr AD |
| 19 | CO01TUNG | iso2k | Coral | -5.2170 | 145.8170 | -3.0 | d18O | d18O | [-4.827, -4.786, -4.693, -4.852, -4.991, -4.90... | permil | d18O | None | subannual | temperature | 1.0 | Ocean2kHR_140_iso2k | year | year | [1993.042, 1992.792, 1992.542, 1992.292, 1992.... | yr AD |
df_iso2k_filt.info()
<class 'pandas.core.frame.DataFrame'>
Index: 49 entries, 2 to 155
Data columns (total 20 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 dataSetName 49 non-null object
1 compilationName 49 non-null object
2 archiveType 49 non-null object
3 geo_meanLat 49 non-null float64
4 geo_meanLon 49 non-null float64
5 geo_meanElev 49 non-null float64
6 paleoData_variableName 49 non-null object
7 paleoData_standardName 49 non-null object
8 paleoData_values 49 non-null object
9 paleoData_units 49 non-null object
10 paleoData_proxy 49 non-null object
11 paleoData_proxyGeneral 0 non-null object
12 paleoData_seasonality 39 non-null object
13 paleoData_interpName 49 non-null object
14 paleoData_interpRank 24 non-null float64
15 TSID 49 non-null object
16 time_variableName 49 non-null object
17 time_standardName 49 non-null object
18 time_values 49 non-null object
19 time_units 49 non-null object
dtypes: float64(4), object(16)
memory usage: 8.0+ KB
We have 49 entries left in the iso2k database.
CoralHydro2k#
Let’s repeat our filtering. The CoralHydro2k database does not have a property indicating whether to use in temperature analysis.
D_coral2k = LiPD()
D_coral2k.load_from_dir('./data/coral2k')
Loading 179 LiPD files
100%|██████████| 179/179 [00:02<00:00, 88.65it/s]
Loaded..
query_coral = """ PREFIX le: <http://linked.earth/ontology#>
PREFIX wgs84: <http://www.w3.org/2003/01/geo/wgs84_pos#>
PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>
SELECT ?dataSetName ?compilationName ?archiveType ?geo_meanLat ?geo_meanLon ?geo_meanElev
?paleoData_variableName ?paleoData_standardName ?paleoData_values ?paleoData_units
?paleoData_proxy ?paleoData_proxyGeneral ?paleoData_seasonality ?paleoData_interpName ?paleoData_interpRank
?TSID ?time_variableName ?time_standardName ?time_values
?time_units where{
?ds a le:Dataset .
?ds le:hasName ?dataSetName .
# Get the archive
OPTIONAL{
?ds le:hasArchiveType ?archiveTypeObj .
?archiveTypeObj rdfs:label ?archiveType .
}
# Geographical information
?ds le:hasLocation ?loc .
OPTIONAL{?loc wgs84:lat ?geo_meanLat .}
OPTIONAL{?loc wgs84:long ?geo_meanLon .}
OPTIONAL{?loc wgs84:alt ?geo_meanElev .}
# PaleoData
?ds le:hasPaleoData ?data .
?data le:hasMeasurementTable ?table .
?table le:hasVariable ?var .
# Name
?var le:hasName ?paleoData_variableName .
?var le:hasStandardVariable ?variable_obj .
?variable_obj rdfs:label ?paleoData_standardName .
#Values
?var le:hasValues ?paleoData_values .
#Seasonality
OPTIONAL{?var le:seasonality ?paleoData_seasonality} .
#Units
OPTIONAL{
?var le:hasUnits ?paleoData_unitsObj .
?paleoData_unitsObj rdfs:label ?paleoData_units .
}
#Proxy information
OPTIONAL{
?var le:hasProxy ?paleoData_proxyObj .
?paleoData_proxyObj rdfs:label ?paleoData_proxy .
}
OPTIONAL{
?var le:hasProxyGeneral ?paleoData_proxyGeneralObj .
?paleoData_proxyGeneralObj rdfs:label ?paleoData_proxyGeneral .
}
# Compilation
?var le:partOfCompilation ?compilation .
?compilation le:hasName ?compilationName .
FILTER (?compilationName = "CoralHydro2k").
# TSiD (should all have them)
OPTIONAL{
?var le:hasVariableId ?TSID
} .
# Interpretation (might be an optional field)
OPTIONAL{
?var le:hasInterpretation ?interp .
?interp le:hasVariable ?interpvar .
BIND(REPLACE(STR(?interpvar), "http://linked.earth/ontology/interpretation#", "") AS ?interpretedVariable_Fallback)
OPTIONAL { ?interpvar rdfs:label ?interpretedVariable_Label } # Use a temporary variable for the label
BIND(COALESCE(?interpretedVariable_Label, ?interpretedVariable_Fallback) AS ?paleoData_interpName) # COALESCE into the final variable
}
OPTIONAL{
?var le:hasInterpretation ?interp .
?interp le:hasRank ?paleoData_interpRank .}
OPTIONAL{
?var le:hasInterpretation ?interp .
?interp le:hasSeasonality ?seasonalityURI .
BIND(REPLACE(STR(?seasonalityURI), "http://linked.earth/ontology/interpretation#", "") AS ?seasonality_Fallback)
OPTIONAL { ?seasonalityURI rdfs:label ?seasonalityLabel }
BIND(COALESCE(?seasonalityLabel, ?seasonality_Fallback) AS ?paleoData_seasonality)
}
#Time information
?table le:hasVariable ?timevar .
?timevar le:hasName ?time_variableName .
?timevar le:hasStandardVariable ?timevar_obj .
?timevar_obj rdfs:label ?time_standardName .
VALUES ?time_standardName {"year"} .
?timevar le:hasValues ?time_values .
OPTIONAL{
?timevar le:hasUnits ?time_unitsObj .
?time_unitsObj rdfs:label ?time_units .
}
}"""
qres, df_coral2k = D_coral2k.query(query_coral)
df_coral2k.head()
| dataSetName | compilationName | archiveType | geo_meanLat | geo_meanLon | geo_meanElev | paleoData_variableName | paleoData_standardName | paleoData_values | paleoData_units | paleoData_proxy | paleoData_proxyGeneral | paleoData_seasonality | paleoData_interpName | paleoData_interpRank | TSID | time_variableName | time_standardName | time_values | time_units | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | KA17RYU01 | CoralHydro2k | Coral | 28.300 | 130.000 | -3.5 | SrCa | Sr/Ca | [8.802, 9.472, 8.825, 9.355, 8.952, 9.297, 8.8... | mmol/mol | None | None | None | None | None | KA17RYU01_SrCa | year | year | [1578.58, 1579.08, 1579.58, 1580.08, 1580.58, ... | yr AD |
| 1 | CH18YOA02 | CoralHydro2k | Coral | 16.448 | 111.605 | NA | SrCa | Sr/Ca | [8.58, 8.683, 8.609, 8.37, 8.38, 8.417, 8.584,... | mmol/mol | None | None | None | None | None | CH18YOA02_SrCa | year | year | [1987.92, 1988.085, 1988.25, 1988.42, 1988.585... | yr AD |
| 2 | FL17DTO02 | CoralHydro2k | Coral | 24.699 | -82.799 | -3.0 | SrCa | Sr/Ca | [9.159, 9.257, 9.245, 9.166, 9.045, 9.013, 8.9... | mmol/mol | None | None | None | None | None | FL17DTO02_SrCa | year | year | [1837.04, 1837.13, 1837.21, 1837.29, 1837.38, ... | yr AD |
| 3 | BO14HTI01 | CoralHydro2k | Coral | 12.210 | 109.310 | -3.6 | d18O | d18O | [-5.4206, -5.3477, -5.1354, -5.7119, -5.9058, ... | permil | None | None | None | None | None | BO14HTI01_d18O | year | year | [1977.37, 1977.45, 1977.54, 1977.62, 1977.7, 1... | yr AD |
| 4 | BO14HTI01 | CoralHydro2k | Coral | 12.210 | 109.310 | -3.6 | SrCa | Sr/Ca | [9.2, 9.17, 9.11, 9.02, 8.95, 8.99, 9.06, 9.1,... | mmol/mol | None | None | None | None | None | BO14HTI01_SrCa | year | year | [1600.04, 1600.12, 1600.2, 1600.28, 1600.37, 1... | yr AD |
Since there is no interpretation objects to go by, let’s have a look at the names of the variables:
df_coral2k['paleoData_variableName'].unique()
array(['SrCa', 'd18O', 'd18O_sw'], dtype=object)
Sr/Ca is usually interpreted as a temperature proxy. d18O contains information about both temperature and d18Osw. Let’s keep these two and drop any duplicates as identified by TSiDs:
df_coral2k_filt = df_coral2k[df_coral2k['paleoData_variableName'].isin(['SrCa', 'd18O'])].drop_duplicates(subset='TSID', keep='first')
df_coral2k_filt.head()
| dataSetName | compilationName | archiveType | geo_meanLat | geo_meanLon | geo_meanElev | paleoData_variableName | paleoData_standardName | paleoData_values | paleoData_units | paleoData_proxy | paleoData_proxyGeneral | paleoData_seasonality | paleoData_interpName | paleoData_interpRank | TSID | time_variableName | time_standardName | time_values | time_units | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | KA17RYU01 | CoralHydro2k | Coral | 28.300 | 130.000 | -3.5 | SrCa | Sr/Ca | [8.802, 9.472, 8.825, 9.355, 8.952, 9.297, 8.8... | mmol/mol | None | None | None | None | None | KA17RYU01_SrCa | year | year | [1578.58, 1579.08, 1579.58, 1580.08, 1580.58, ... | yr AD |
| 1 | CH18YOA02 | CoralHydro2k | Coral | 16.448 | 111.605 | NA | SrCa | Sr/Ca | [8.58, 8.683, 8.609, 8.37, 8.38, 8.417, 8.584,... | mmol/mol | None | None | None | None | None | CH18YOA02_SrCa | year | year | [1987.92, 1988.085, 1988.25, 1988.42, 1988.585... | yr AD |
| 2 | FL17DTO02 | CoralHydro2k | Coral | 24.699 | -82.799 | -3.0 | SrCa | Sr/Ca | [9.159, 9.257, 9.245, 9.166, 9.045, 9.013, 8.9... | mmol/mol | None | None | None | None | None | FL17DTO02_SrCa | year | year | [1837.04, 1837.13, 1837.21, 1837.29, 1837.38, ... | yr AD |
| 3 | BO14HTI01 | CoralHydro2k | Coral | 12.210 | 109.310 | -3.6 | d18O | d18O | [-5.4206, -5.3477, -5.1354, -5.7119, -5.9058, ... | permil | None | None | None | None | None | BO14HTI01_d18O | year | year | [1977.37, 1977.45, 1977.54, 1977.62, 1977.7, 1... | yr AD |
| 4 | BO14HTI01 | CoralHydro2k | Coral | 12.210 | 109.310 | -3.6 | SrCa | Sr/Ca | [9.2, 9.17, 9.11, 9.02, 8.95, 8.99, 9.06, 9.1,... | mmol/mol | None | None | None | None | None | BO14HTI01_SrCa | year | year | [1600.04, 1600.12, 1600.2, 1600.28, 1600.37, 1... | yr AD |
df_coral2k_filt.info()
<class 'pandas.core.frame.DataFrame'>
Index: 233 entries, 0 to 251
Data columns (total 20 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 dataSetName 233 non-null object
1 compilationName 233 non-null object
2 archiveType 233 non-null object
3 geo_meanLat 233 non-null float64
4 geo_meanLon 233 non-null float64
5 geo_meanElev 232 non-null object
6 paleoData_variableName 233 non-null object
7 paleoData_standardName 233 non-null object
8 paleoData_values 233 non-null object
9 paleoData_units 233 non-null object
10 paleoData_proxy 0 non-null object
11 paleoData_proxyGeneral 0 non-null object
12 paleoData_seasonality 0 non-null object
13 paleoData_interpName 0 non-null object
14 paleoData_interpRank 0 non-null object
15 TSID 233 non-null object
16 time_variableName 233 non-null object
17 time_standardName 233 non-null object
18 time_values 233 non-null object
19 time_units 233 non-null object
dtypes: float64(2), object(18)
memory usage: 38.2+ KB
This database adds 233 new records.
DataBase: Assemble!#
Let’s concatenate the three DataFrames into a clean database, dropping timeseries with the same TSiD as we go. Let’s also convert the values to numpy.array:
df_raw = pd.concat([df_pages2k, df_iso2k_filt, df_coral2k_filt], ignore_index=True).drop_duplicates(subset='TSID', keep='first')
df_raw['paleoData_values']=df_raw['paleoData_values'].apply(lambda row : json.loads(row) if isinstance(row, str) else row)
df_raw['time_values']=df_raw['time_values'].apply(lambda row : json.loads(row) if isinstance(row, str) else row)
df_raw.info()
<class 'pandas.core.frame.DataFrame'>
Index: 972 entries, 0 to 972
Data columns (total 20 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 dataSetName 972 non-null object
1 compilationName 972 non-null object
2 archiveType 972 non-null object
3 geo_meanLat 972 non-null float64
4 geo_meanLon 972 non-null float64
5 geo_meanElev 964 non-null object
6 paleoData_variableName 972 non-null object
7 paleoData_standardName 972 non-null object
8 paleoData_values 972 non-null object
9 paleoData_units 564 non-null object
10 paleoData_proxy 739 non-null object
11 paleoData_proxyGeneral 500 non-null object
12 paleoData_seasonality 729 non-null object
13 paleoData_interpName 739 non-null object
14 paleoData_interpRank 24 non-null float64
15 TSID 972 non-null object
16 time_variableName 972 non-null object
17 time_standardName 972 non-null object
18 time_values 972 non-null object
19 time_units 972 non-null object
dtypes: float64(3), object(17)
memory usage: 159.5+ KB
We have 972 timeseries before any further pre-processing.
Special handling: The Palmyra record#
The Palmyra Coral record in these databases may not have been updated with the latest changes published by Dee et al. (2020). Let’s open this record and extract the same information:
palm = LiPD()
palm.load('./data/Palmyra.Dee.2020.lpd')
Loading 1 LiPD files
100%|██████████| 1/1 [00:00<00:00, 9.18it/s]
Loaded..
query_palm = """
PREFIX le: <http://linked.earth/ontology#>
PREFIX wgs84: <http://www.w3.org/2003/01/geo/wgs84_pos#>
PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>
SELECT ?dataSetName ?compilationName ?archiveType ?geo_meanLat ?geo_meanLon ?geo_meanElev
?paleoData_variableName ?paleoData_standardName ?paleoData_values ?paleoData_units
?paleoData_proxy ?paleoData_proxyGeneral ?paleoData_seasonality ?paleoData_interpName ?paleoData_interpRank
?TSID ?time_variableName ?time_standardName ?time_values
?time_units where{
?ds a le:Dataset .
?ds le:hasName ?dataSetName .
# Get the archive
OPTIONAL{
?ds le:hasArchiveType ?archiveTypeObj .
?archiveTypeObj rdfs:label ?archiveType .
}
# Geographical information
?ds le:hasLocation ?loc .
OPTIONAL{?loc wgs84:lat ?geo_meanLat .}
OPTIONAL{?loc wgs84:long ?geo_meanLon .}
OPTIONAL{?loc wgs84:alt ?geo_meanElev .}
# PaleoData
?ds le:hasPaleoData ?data .
?data le:hasMeasurementTable ?table .
?table le:hasVariable ?var .
# Name
?var le:hasName ?paleoData_variableName .
?var le:hasStandardVariable ?variable_obj .
?variable_obj rdfs:label ?paleoData_standardName .
#Values
?var le:hasValues ?paleoData_values .
#Units
OPTIONAL{
?var le:hasUnits ?paleoData_unitsObj .
?paleoData_unitsObj rdfs:label ?paleoData_units .
}
#Proxy information
OPTIONAL{
?var le:hasProxy ?paleoData_proxyObj .
?paleoData_proxyObj rdfs:label ?paleoData_proxy .
}
OPTIONAL{
?var le:hasProxyGeneral ?paleoData_proxyGeneralObj .
?paleoData_proxyGeneralObj rdfs:label ?paleoData_proxyGeneral .
}
# Compilation
OPTIONAL{
?var le:partOfCompilation ?compilation .
?compilation le:hasName ?compilationName .}
# TSiD (should all have them)
OPTIONAL{
?var le:hasVariableId ?TSID
} .
# Interpretation (might be an optional field)
OPTIONAL{
?var le:hasInterpretation ?interp .
?interp le:hasVariable ?interpvar .
BIND(REPLACE(STR(?interpvar), "http://linked.earth/ontology/interpretation#", "") AS ?interpretedVariable_Fallback)
OPTIONAL { ?interpvar rdfs:label ?interpretedVariable_Label } # Use a temporary variable for the label
BIND(COALESCE(?interpretedVariable_Label, ?interpretedVariable_Fallback) AS ?paleoData_interpName) # COALESCE into the final variable
}
OPTIONAL{
?var le:hasInterpretation ?interp .
?interp le:hasRank ?paleoData_interpRank .}
OPTIONAL{
?var le:hasInterpretation ?interp .
?interp le:hasSeasonality ?seasonalityURI .
BIND(REPLACE(STR(?seasonalityURI), "http://linked.earth/ontology/interpretation#", "") AS ?seasonality_Fallback)
OPTIONAL { ?seasonalityURI rdfs:label ?seasonalityLabel }
BIND(COALESCE(?seasonalityLabel, ?seasonality_Fallback) AS ?paleoData_seasonality)
}
#Time information
?table le:hasVariable ?timevar .
?timevar le:hasName ?time_variableName .
?timevar le:hasStandardVariable ?timevar_obj .
?timevar_obj rdfs:label ?time_standardName .
VALUES ?time_standardName {"year"} .
?timevar le:hasValues ?time_values .
OPTIONAL{
?timevar le:hasUnits ?time_unitsObj .
?time_unitsObj rdfs:label ?time_units .
}
}"""
qres, df_palm = palm.query(query_palm)
df_palm.head()
| dataSetName | compilationName | archiveType | geo_meanLat | geo_meanLon | geo_meanElev | paleoData_variableName | paleoData_standardName | paleoData_values | paleoData_units | paleoData_proxy | paleoData_proxyGeneral | paleoData_seasonality | paleoData_interpName | paleoData_interpRank | TSID | time_variableName | time_standardName | time_values | time_units | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Palmyra.Dee.2020 | None | Coral | 5.8664 | -162.12 | -9.0 | d18O | d18O | [-4.749, -4.672, -4.724, -4.717, -4.8947, -4.8... | permil | d18O | isotopic | subannual | seawaterIsotope | 1.0 | PCU-1cce3af5639a4ec | year | year | [1146.375, 1146.4583, 1146.5417, 1146.625, 114... | yr AD |
| 1 | Palmyra.Dee.2020 | None | Coral | 5.8664 | -162.12 | -9.0 | d18O | d18O | [-4.749, -4.672, -4.724, -4.717, -4.8947, -4.8... | permil | d18O | isotopic | subannual | temperature | NaN | PCU-1cce3af5639a4ec | year | year | [1146.375, 1146.4583, 1146.5417, 1146.625, 114... | yr AD |
| 2 | Palmyra.Dee.2020 | None | Coral | 5.8664 | -162.12 | -9.0 | year | year | [1146.375, 1146.4583, 1146.5417, 1146.625, 114... | yr AD | None | None | None | None | NaN | PCU-45b2ff49e3944f1 | year | year | [1146.375, 1146.4583, 1146.5417, 1146.625, 114... | yr AD |
Let’s keep the variable associated with the temperature interpretation:
df_palm_filt = df_palm[df_palm['paleoData_interpName']=='temperature']
df_palm_filt
| dataSetName | compilationName | archiveType | geo_meanLat | geo_meanLon | geo_meanElev | paleoData_variableName | paleoData_standardName | paleoData_values | paleoData_units | paleoData_proxy | paleoData_proxyGeneral | paleoData_seasonality | paleoData_interpName | paleoData_interpRank | TSID | time_variableName | time_standardName | time_values | time_units | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Palmyra.Dee.2020 | None | Coral | 5.8664 | -162.12 | -9.0 | d18O | d18O | [-4.749, -4.672, -4.724, -4.717, -4.8947, -4.8... | permil | d18O | isotopic | subannual | temperature | NaN | PCU-1cce3af5639a4ec | year | year | [1146.375, 1146.4583, 1146.5417, 1146.625, 114... | yr AD |
Let’s make sure that all the values are converted from string:
df_palm_filt['paleoData_values']=df_palm_filt['paleoData_values'].apply(lambda row : json.loads(row) if isinstance(row, str) else row)
df_palm_filt['time_values']=df_palm_filt['time_values'].apply(lambda row : json.loads(row) if isinstance(row, str) else row)
Handling duplication: Sr/Ca#
Now, let’s find the entry(ies) within PAGES2k corresponding to the Palmyra record. To do so, let’s find all the records within 50km:
# Earth's radius in kilometers
R = 6371.0
# Threshold
threshold = 50 #Look for all records within threshold km
# Target location
target_lat = df_palm_filt['geo_meanLat'].iloc[0]
target_lon = df_palm_filt['geo_meanLon'].iloc[0]
# Convert degrees to radians
lat1 = np.radians(target_lat)
lon1 = np.radians(target_lon)
lat2 = np.radians(df_raw['geo_meanLat'])
lon2 = np.radians(df_raw['geo_meanLon'])
# Haversine formula
dlat = lat2 - lat1
dlon = lon2 - lon1
a = np.sin(dlat / 2)**2 + np.cos(lat1) * np.cos(lat2) * np.sin(dlon / 2)**2
c = 2 * np.arcsin(np.sqrt(a))
distance_km = R * c
# Add distance column
df_raw['distance_km'] = distance_km
# Filter to only rows within 50 km
df_distance = df_raw[df_raw['distance_km'] <= threshold].reset_index(drop=True)
df_distance
| dataSetName | compilationName | archiveType | geo_meanLat | geo_meanLon | geo_meanElev | paleoData_variableName | paleoData_standardName | paleoData_values | paleoData_units | ... | paleoData_proxyGeneral | paleoData_seasonality | paleoData_interpName | paleoData_interpRank | TSID | time_variableName | time_standardName | time_values | time_units | distance_km | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Ocn-Palmyra.Nurhati.2011 | Pages2kTemperature | Coral | 5.870 | -162.130 | -10.0 | Sr/Ca | Sr/Ca | [8.96, 8.9, 8.91, 8.94, 8.92, 8.89, 8.87, 8.81... | mmol/mol | ... | None | subannual | temperature | NaN | Ocean2kHR_161 | year | year | [1998.29, 1998.21, 1998.13, 1998.04, 1997.96, ... | yr AD | 1.176328 |
| 1 | Ocn-Palmyra.Cobb.2013 | Pages2kTemperature | Coral | 5.870 | -162.130 | -7.0 | d18O | d18O | [-4.651, -4.631, -4.629, -4.562, -4.58, -4.442... | permil | ... | None | subannual | temperature | NaN | Ocean2kHR_139 | year | year | [928.125, 928.209, 928.292, 928.375, 928.459, ... | yr AD | 1.176328 |
| 2 | CO03COPM | iso2k | Coral | 5.870 | -162.130 | -7.0 | d18O | d18O | [-5.27, -5.42, -5.49, -5.5, -5.44, -5.48, -5.5... | permil | ... | None | subannual | temperature | NaN | Ocean2kHR_139_iso2k | year | year | [1998.37, 1998.29, 1998.2, 1998.13, 1998.04, 1... | yr AD | 1.176328 |
| 3 | NU11PAL01 | CoralHydro2k | Coral | 5.867 | -162.133 | -9.0 | d18O | d18O | [-4.79, -4.89, -4.81, -4.84, -4.85, -4.82, -4.... | permil | ... | None | None | None | NaN | NU11PAL01_d18O | year | year | [1886.13, 1886.21, 1886.29, 1886.38, 1886.46, ... | yr AD | 1.439510 |
| 4 | NU11PAL01 | CoralHydro2k | Coral | 5.867 | -162.133 | -9.0 | SrCa | Sr/Ca | [9.01, 9.02, 8.98, 8.99, 9.0, 8.99, 9.0, 9.11,... | mmol/mol | ... | None | None | None | NaN | NU11PAL01_SrCa | year | year | [1886.13, 1886.21, 1886.29, 1886.38, 1886.46, ... | yr AD | 1.439510 |
| 5 | SA19PAL01 | CoralHydro2k | Coral | 5.878 | -162.142 | -10.0 | d18O | d18O | [-5.21, -5.23, -5.17, -5.13, -5.11, -5.05, -5.... | permil | ... | None | None | None | NaN | SA19PAL01_d18O | year | year | [1983.58, 1983.67, 1983.75, 1983.83, 1983.92, ... | yr AD | 2.754166 |
| 6 | SA19PAL01 | CoralHydro2k | Coral | 5.878 | -162.142 | -10.0 | SrCa | Sr/Ca | [8.94, 9.08, 9.17, 9.16, 9.16, 9.17, 9.17, 9.1... | mmol/mol | ... | None | None | None | NaN | SA19PAL01_SrCa | year | year | [1980.75, 1980.83, 1980.92, 1981.0, 1981.08, 1... | yr AD | 2.754166 |
| 7 | SA19PAL02 | CoralHydro2k | Coral | 5.878 | -162.142 | -10.0 | d18O | d18O | [-5.05, -5.1, -5.21, -5.14, -5.23, -5.32, -5.3... | permil | ... | None | None | None | NaN | SA19PAL02_d18O | year | year | [1985.92, 1986.0, 1986.08, 1986.17, 1986.25, 1... | yr AD | 2.754166 |
| 8 | SA19PAL02 | CoralHydro2k | Coral | 5.878 | -162.142 | -10.0 | SrCa | Sr/Ca | [8.86, 8.82, 8.81, 8.85, 8.89, 8.87, 8.82, 8.8... | mmol/mol | ... | None | None | None | NaN | SA19PAL02_SrCa | year | year | [1985.33, 1985.42, 1985.5, 1985.58, 1985.67, 1... | yr AD | 2.754166 |
| 9 | CO03PAL08 | CoralHydro2k | Coral | 5.870 | -162.130 | NA | d18O | d18O | [-5.14, -5.13, -5.13, -5.16, -5.28, -4.99, -4.... | permil | ... | None | None | None | NaN | CO03PAL08_d18O | year | year | [1653.63, 1653.65, 1653.686083, 1653.722167, 1... | yr AD | 1.176328 |
| 10 | CO03PAL09 | CoralHydro2k | Coral | 5.870 | -162.130 | NA | d18O | d18O | [-4.98, -4.8, -4.68, -4.74, -4.7, -4.74, -4.84... | permil | ... | None | None | None | NaN | CO03PAL09_d18O | year | year | [1664.96, 1665.0215, 1665.083, 1665.164, 1665.... | yr AD | 1.176328 |
| 11 | CO03PAL04 | CoralHydro2k | Coral | 5.870 | -162.130 | NA | d18O | d18O | [-4.86, -4.9, -5.0, -4.92, -4.91, -4.85, -4.74... | permil | ... | None | None | None | NaN | CO03PAL04_d18O | year | year | [1398.68, 1398.747167, 1398.814333, 1398.8815,... | yr AD | 1.176328 |
| 12 | CO03PAL10 | CoralHydro2k | Coral | 5.870 | -162.130 | NA | d18O | d18O | [-4.865, -4.765, -4.805, -4.665, -4.595, -4.73... | permil | ... | None | None | None | NaN | CO03PAL10_d18O | year | year | [1326.65, 1326.75825, 1326.8665, 1326.97475, 1... | yr AD | 1.176328 |
| 13 | CO03PAL05 | CoralHydro2k | Coral | 5.870 | -162.130 | NA | d18O | d18O | [-4.61, -4.64, -4.7, -4.77, -4.86, -4.85, -4.8... | permil | ... | None | None | None | NaN | CO03PAL05_d18O | year | year | [1405.1569, 1405.280175, 1405.40345, 1405.5267... | yr AD | 1.176328 |
| 14 | CO03PAL07 | CoralHydro2k | Coral | 5.870 | -162.130 | NA | d18O | d18O | [-4.63, -4.55, -4.65, -4.59, -4.74, -4.79, -4.... | permil | ... | None | None | None | NaN | CO03PAL07_d18O | year | year | [1635.02, 1635.083, 1635.146, 1635.209, 1635.2... | yr AD | 1.176328 |
| 15 | CO03PAL06 | CoralHydro2k | Coral | 5.870 | -162.130 | NA | d18O | d18O | [-4.64, -4.64, -4.62, -4.62, -4.68, -4.77, -4.... | permil | ... | None | None | None | NaN | CO03PAL06_d18O | year | year | [1411.9262, 1412.0046, 1412.083, 1412.1775, 14... | yr AD | 1.176328 |
| 16 | CO03PAL02 | CoralHydro2k | Coral | 5.870 | -162.130 | NA | d18O | d18O | [-4.631, -4.724, -4.709, -4.707, -4.912, -4.83... | permil | ... | None | None | None | NaN | CO03PAL02_d18O | year | year | [1149.08, 1149.2225, 1149.365, 1149.5075, 1149... | yr AD | 1.176328 |
| 17 | CO03PAL03 | CoralHydro2k | Coral | 5.870 | -162.130 | NA | d18O | d18O | [-4.79, -4.73, -4.66, -4.66, -4.78, -4.69, -4.... | permil | ... | None | None | None | NaN | CO03PAL03_d18O | year | year | [1317.17, 1317.29, 1317.41, 1317.53, 1317.65, ... | yr AD | 1.176328 |
| 18 | CO03PAL01 | CoralHydro2k | Coral | 5.870 | -162.130 | NA | d18O | d18O | [-4.68, -4.62, -4.65, -4.56, -4.58, -4.42, -4.... | permil | ... | None | None | None | NaN | CO03PAL01_d18O | year | year | [928.08, 928.175, 928.27, 928.365, 928.46, 928... | yr AD | 1.176328 |
19 rows × 21 columns
The first 18 rows seem to be corresponding to various coral heads within the Palmyra records. Let’s have a look at them. Let’s first have a look at Sr/Ca record:
df_distance_SrCa = df_distance[df_distance['paleoData_variableName'].isin(['SrCa', 'Sr/Ca'])]
df_distance_SrCa
| dataSetName | compilationName | archiveType | geo_meanLat | geo_meanLon | geo_meanElev | paleoData_variableName | paleoData_standardName | paleoData_values | paleoData_units | ... | paleoData_proxyGeneral | paleoData_seasonality | paleoData_interpName | paleoData_interpRank | TSID | time_variableName | time_standardName | time_values | time_units | distance_km | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Ocn-Palmyra.Nurhati.2011 | Pages2kTemperature | Coral | 5.870 | -162.130 | -10.0 | Sr/Ca | Sr/Ca | [8.96, 8.9, 8.91, 8.94, 8.92, 8.89, 8.87, 8.81... | mmol/mol | ... | None | subannual | temperature | NaN | Ocean2kHR_161 | year | year | [1998.29, 1998.21, 1998.13, 1998.04, 1997.96, ... | yr AD | 1.176328 |
| 4 | NU11PAL01 | CoralHydro2k | Coral | 5.867 | -162.133 | -9.0 | SrCa | Sr/Ca | [9.01, 9.02, 8.98, 8.99, 9.0, 8.99, 9.0, 9.11,... | mmol/mol | ... | None | None | None | NaN | NU11PAL01_SrCa | year | year | [1886.13, 1886.21, 1886.29, 1886.38, 1886.46, ... | yr AD | 1.439510 |
| 6 | SA19PAL01 | CoralHydro2k | Coral | 5.878 | -162.142 | -10.0 | SrCa | Sr/Ca | [8.94, 9.08, 9.17, 9.16, 9.16, 9.17, 9.17, 9.1... | mmol/mol | ... | None | None | None | NaN | SA19PAL01_SrCa | year | year | [1980.75, 1980.83, 1980.92, 1981.0, 1981.08, 1... | yr AD | 2.754166 |
| 8 | SA19PAL02 | CoralHydro2k | Coral | 5.878 | -162.142 | -10.0 | SrCa | Sr/Ca | [8.86, 8.82, 8.81, 8.85, 8.89, 8.87, 8.82, 8.8... | mmol/mol | ... | None | None | None | NaN | SA19PAL02_SrCa | year | year | [1985.33, 1985.42, 1985.5, 1985.58, 1985.67, 1... | yr AD | 2.754166 |
4 rows × 21 columns
Let’s plot them:
ts_list = []
for _, row in df_distance_SrCa.iterrows():
proxy = row['paleoData_proxy'] if isinstance(row['paleoData_proxy'],str) else row['paleoData_variableName']
ts_list.append(pyleo.GeoSeries(time=row['time_values'],value=row['paleoData_values'],
time_name='Time',value_name=row['paleoData_variableName'],
time_unit=row['time_units'], value_unit=row['paleoData_units'],
lat = row['geo_meanLat'], lon = row['geo_meanLon'],
elevation = row['geo_meanElev'], importedFrom='lipdverse.org',
archiveType = row['archiveType'],
observationType=row['paleoData_proxy'],
label=row['dataSetName']+'_'+proxy, verbose = False))
mgs_srca = pyleo.MultipleGeoSeries(ts_list)
fig, ax = mgs_srca.stackplot()
fig.show()
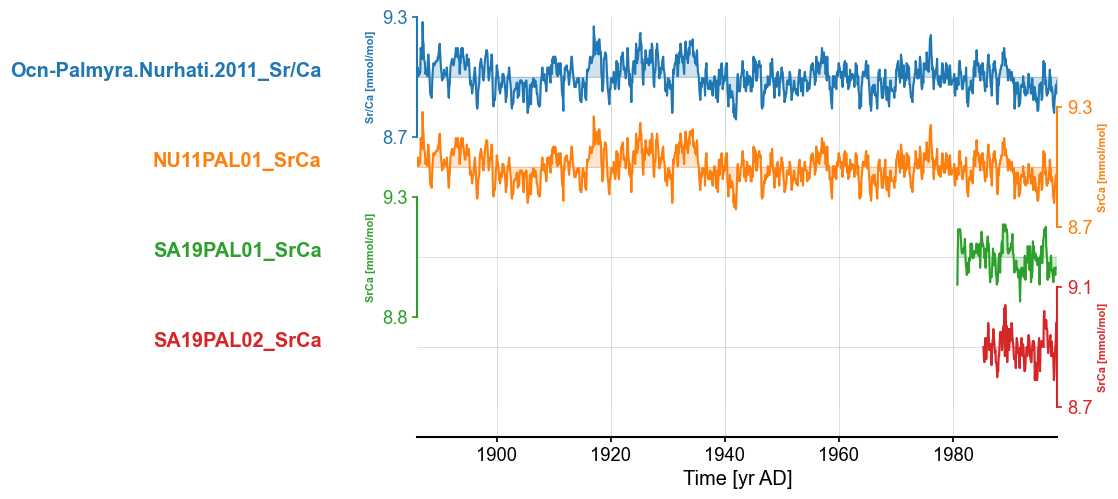
The three records look very similar. The Ocn-Palmyra.Nurhati.2011_Sr/Ca and NU11PAL01_SrCa appear nearly identical. However, this may occur in other places in the database, so we want an algorithm to (1) find records that may be the same. We can follow a similar strategy as we have done here. a. Are they within 50km of each other and b. do they have the same or similar variable names, and (2) assess whether these records are, in fact, the same.
We can do this in several ways:
Correlation: this may work well for true duplicates but may fail for composites
Dynamic Time Warping (DTW), which can be used to to assess shape similarity even when time points aren’t exactly aligned. DTW returns a distance metric: lower values mean more similar patterns.It can be combined with a z-normalization to remove mean/variance effects.
Principal Component Analysis (PCA)/ On groups of nearby records, run PCA to detect if certain records contribute nearly identical structure to the same components. High loadings on the same component can suggest redundancy.
Let’s have a look at these three options.
Option 1: Correlation#
Let’s use the Pyleoclim correlation function. The following code is inspired from this tutorial.
n = len(mgs_srca.series_list)
corr_mat = np.empty((n,n))
corr_mat[:] = np.nan #let's only work on half of the matrix to speeds things up
for i in range(0, n-1): # loop over records
ts = mgs_srca.series_list[i]
mgsl = pyleo.MultipleGeoSeries(mgs_srca.series_list[i+1:]) #gather potential pairs
corr = mgsl.correlation(ts,method='built-in') # compute correlations
corr_mat[i,i+1:] = corr.r # save result in matrix
Looping over 3 Series in collection
100%|██████████| 3/3 [00:00<00:00, 193.66it/s]
Looping over 2 Series in collection
100%|██████████| 2/2 [00:00<00:00, 288.76it/s]
Looping over 1 Series in collection
100%|██████████| 1/1 [00:00<00:00, 336.57it/s]
labels = []
for item in mgs_srca.series_list:
labels.append(item.label)
core_matrix_df = pd.DataFrame(corr_mat, index=labels, columns=labels)
plt.figure(figsize=(10, 8))
sns.heatmap(core_matrix_df, annot=True, fmt=".2f", cmap="coolwarm", center=0)
plt.title("Correlation Heatmap")
plt.tight_layout()
plt.show()
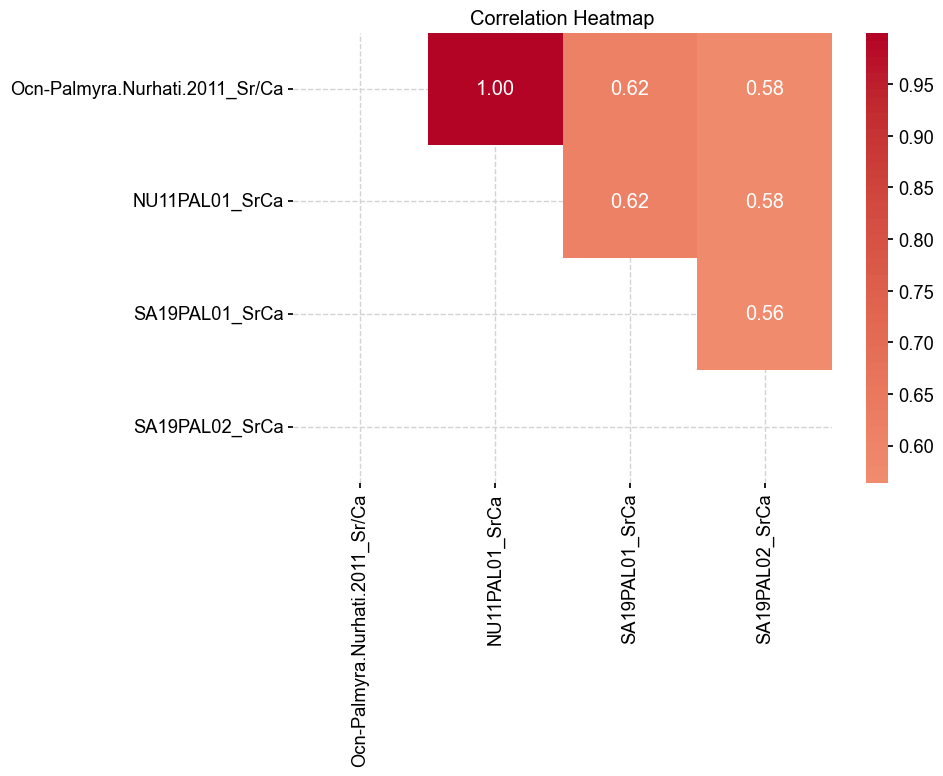
Unsurprisingly, Ocn-Palmyra.Nurhati.2011_Sr/Ca and NU11PAL01_SrCa have a correlation of 1 as they are essentially the same record. The other correlations are below 0.8, which makes it difficult to assess whether they are, in fact the same record. Let’s have a look at DTW as a potential discriminant.
Option 2: Dynamic Time Wrapping#
Dynamic Time Wrapping (DTW) is a distance metric designed to compare time series that may be misaligned in time or differ in length. Unlike traditional correlation-based methods, DTW allows for flexible, non-linear alignment of sequences by warping the time axis to minimize the total distance between corresponding points. In our context, DTW provides a more robust measure of similarity between records than correlation, especially when records are composites or partially overlapping. By restricting comparisons to shared time intervals and z-score normalizing the data to emphasize shape similarity, DTW helps identify potentially redundant or duplicated records while accounting for the complexities inherent in paleoenvironmental data.
The following cell does the following:
Clip the timeseries for similar time range for comparison
Compute zscore
Compute the DTW distance
# --- Helper function: clip to overlapping time range and normalize ---
def clip_to_overlap(time1, series1, time2, series2, min_overlap=5):
start = max(time1[0], time2[0])
end = min(time1[-1], time2[-1])
mask1 = (time1 >= start) & (time1 <= end)
mask2 = (time2 >= start) & (time2 <= end)
if np.sum(mask1) < min_overlap or np.sum(mask2) < min_overlap:
return None, None # Not enough overlapping points
return zscore(series1[mask1]), zscore(series2[mask2])
# --- Prepare the data ----
records = {}
times = {}
for series in mgs_srca.series_list:
records[series.label] = series.value
times[series.label] = series.time
# --- Main loop ---
labels = list(records.keys())
n = len(labels)
dtw_matrix = np.full((n, n), np.nan)
for i, j in itertools.combinations(range(n), 2):
key_i, key_j = labels[i], labels[j]
s1, s2 = clip_to_overlap(times[key_i], records[key_i],
times[key_j], records[key_j])
if s1 is not None:
path = dtw.warping_path(s1, s2)
dist = dtw.distance(s1, s2)
norm_dist = dist / len(path) # Per-step normalization
dtw_matrix[i, j] = dtw_matrix[j, i] = norm_dist
# Convert to DataFrame for visualization
dtw_df = pd.DataFrame(dtw_matrix, index=labels, columns=labels)
Let’s visualize the results:
plt.figure(figsize=(10, 8))
sns.heatmap(dtw_df, annot=True, fmt=".2f", cmap="viridis", mask=np.isnan(dtw_df))
plt.title("DTW Distance Matrix (Clipped to Overlap)")
plt.tight_layout()
plt.show()
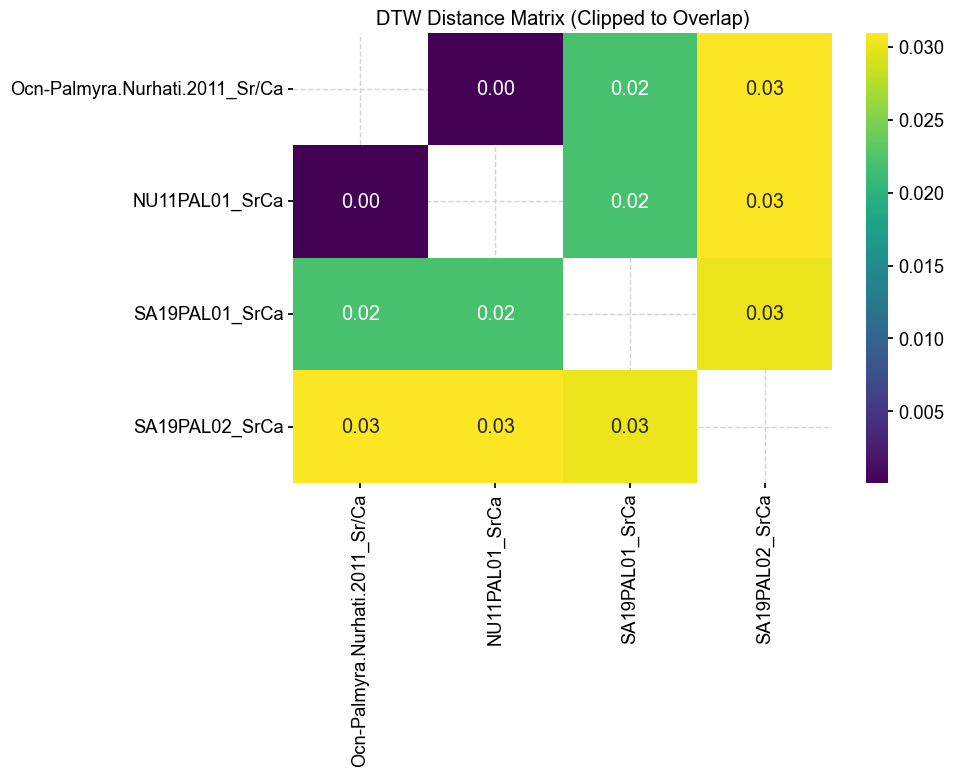
The next problem lies in the interpretation of these results. The matrix holds pairwise DTW distances between time series. Each entry DTW[i, j] is a non-negative real number representing how similar two z-normalized time series are after optimal alignment.
Lower values = more similar shape (after warping).
Higher values = less similar (more misaligned or different structure).
Since we z-scored normalized the data and accounted for number of steps, the DTW distances can be interpreted as follows:
Normalized DTW |
Meaning |
|---|---|
0.0 – 0.1 |
Very similar shape — likely same core or duplicated segment |
0.1 – 0.2 |
High similarity — possibly same site, maybe overlapping or composited |
0.2 – 0.4 |
Moderate similarity — same region, but different signal characteristics |
> 0.4 |
Different shape — likely independent records |
Option 3: Principal Component Analysis#
Let’s leverage the PCA functionality in Pyleoclim. A tutorial for this functionality is available here.
The first step is to brings the records on a common time axis and standardize them:
mgs_srca_sc = mgs_srca.common_time().standardize()
fig, ax = mgs_srca_sc.stackplot()
Let’s run PCA:
pca = mgs_srca_sc.pca()
And let’s have a look at the first mode:
fig, ax = pca.modeplot()
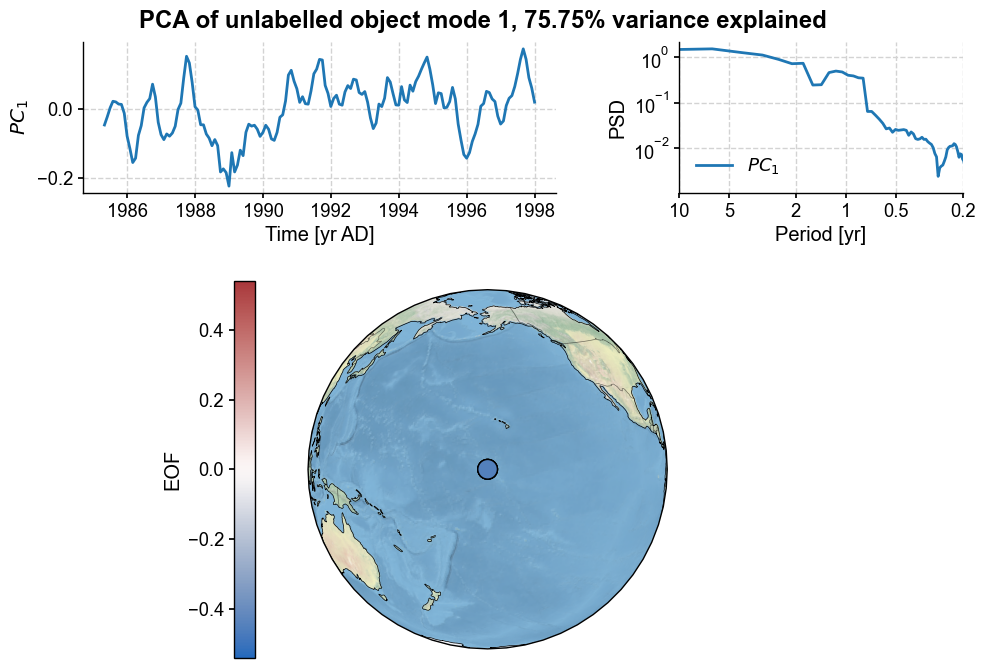
Not unsurprisingly, the first PC explains over 75% of the variance. Let’s have a look at the loads:
pca.eigvecs[1]
array([-0.54194915, 0.4525358 , 0.03876175, -0.70710678])
The loads are not very similar, which is interesting considering that tow of these records are definitely the same. PCA may not work well to find duplicates.
Based on this small proof of concept, correlation and DTW seems to be a reasonable approach to identify duplicates in the database.
Handling duplication part 2: \(\delta^{18}\mathrm{O}\)#
Let’s now have a look at the various \(\delta^{18}\mathrm{O}\) records.
df_distance_d18O = df_distance[df_distance['paleoData_variableName']=='d18O']
df_distance_d18O.head()
| dataSetName | compilationName | archiveType | geo_meanLat | geo_meanLon | geo_meanElev | paleoData_variableName | paleoData_standardName | paleoData_values | paleoData_units | ... | paleoData_proxyGeneral | paleoData_seasonality | paleoData_interpName | paleoData_interpRank | TSID | time_variableName | time_standardName | time_values | time_units | distance_km | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Ocn-Palmyra.Cobb.2013 | Pages2kTemperature | Coral | 5.870 | -162.130 | -7.0 | d18O | d18O | [-4.651, -4.631, -4.629, -4.562, -4.58, -4.442... | permil | ... | None | subannual | temperature | NaN | Ocean2kHR_139 | year | year | [928.125, 928.209, 928.292, 928.375, 928.459, ... | yr AD | 1.176328 |
| 2 | CO03COPM | iso2k | Coral | 5.870 | -162.130 | -7.0 | d18O | d18O | [-5.27, -5.42, -5.49, -5.5, -5.44, -5.48, -5.5... | permil | ... | None | subannual | temperature | NaN | Ocean2kHR_139_iso2k | year | year | [1998.37, 1998.29, 1998.2, 1998.13, 1998.04, 1... | yr AD | 1.176328 |
| 3 | NU11PAL01 | CoralHydro2k | Coral | 5.867 | -162.133 | -9.0 | d18O | d18O | [-4.79, -4.89, -4.81, -4.84, -4.85, -4.82, -4.... | permil | ... | None | None | None | NaN | NU11PAL01_d18O | year | year | [1886.13, 1886.21, 1886.29, 1886.38, 1886.46, ... | yr AD | 1.439510 |
| 5 | SA19PAL01 | CoralHydro2k | Coral | 5.878 | -162.142 | -10.0 | d18O | d18O | [-5.21, -5.23, -5.17, -5.13, -5.11, -5.05, -5.... | permil | ... | None | None | None | NaN | SA19PAL01_d18O | year | year | [1983.58, 1983.67, 1983.75, 1983.83, 1983.92, ... | yr AD | 2.754166 |
| 7 | SA19PAL02 | CoralHydro2k | Coral | 5.878 | -162.142 | -10.0 | d18O | d18O | [-5.05, -5.1, -5.21, -5.14, -5.23, -5.32, -5.3... | permil | ... | None | None | None | NaN | SA19PAL02_d18O | year | year | [1985.92, 1986.0, 1986.08, 1986.17, 1986.25, 1... | yr AD | 2.754166 |
5 rows × 21 columns
ts_list = []
for _, row in df_distance_d18O.iterrows():
proxy = row['paleoData_proxy'] if isinstance(row['paleoData_proxy'],str) else row['paleoData_variableName']
ts_list.append(pyleo.GeoSeries(time=row['time_values'],value=row['paleoData_values'],
time_name='Time',value_name=row['paleoData_variableName'],
time_unit=row['time_units'], value_unit=row['paleoData_units'],
lat = row['geo_meanLat'], lon = row['geo_meanLon'],
elevation = row['geo_meanElev'], importedFrom='lipdverse.org',
archiveType = row['archiveType'],
observationType=row['paleoData_proxy'],
label=row['dataSetName']+'_'+proxy, verbose = False))
# Add the Dee et al. record
ts_list.append(pyleo.GeoSeries(time=df_palm_filt['time_values'].iloc[0],
value=df_palm_filt['paleoData_values'].iloc[0],
time_name='Time',
value_name=df_palm_filt['paleoData_variableName'].iloc[0],
time_unit=df_palm_filt['time_units'].iloc[0],
value_unit=df_palm_filt['paleoData_units'].iloc[0],
lat = df_palm_filt['geo_meanLat'].iloc[0],
lon = df_palm_filt['geo_meanLon'].iloc[0],
elevation = df_palm_filt['geo_meanElev'].iloc[0],
importedFrom='lipdverse.org',
archiveType = df_palm_filt['archiveType'].iloc[0],
observationType=df_palm_filt['paleoData_proxy'].iloc[0],
label=df_palm_filt['dataSetName'].iloc[0]+'_'+proxy, verbose = False))
mgs = pyleo.MultipleGeoSeries(ts_list)
fig, ax = mgs.stackplot()
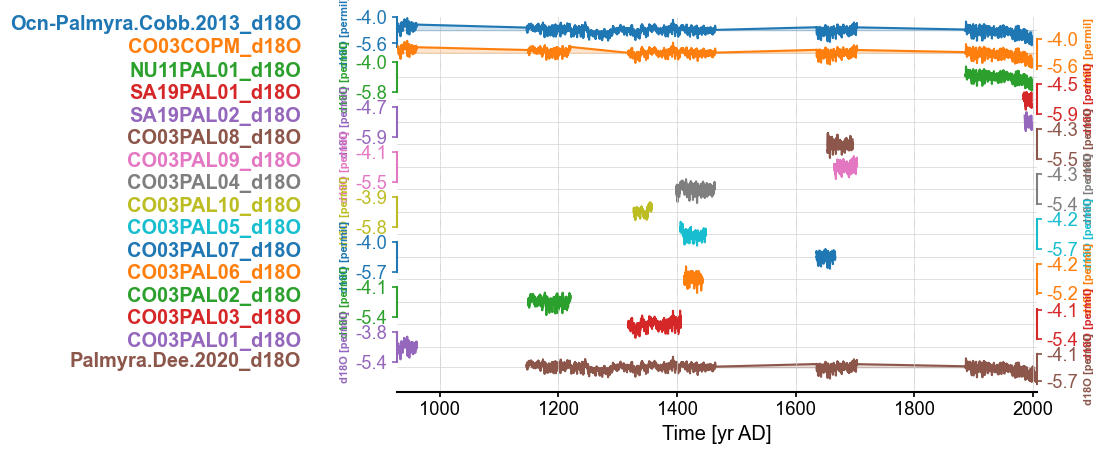
As we have seen with the Sr/Ca record, there seems to be duplications among the record. Although some look like changes in the age models. Let’s explore correlation and DTW to weed out the duplicates.
Option 1: Correlation#
n = len(mgs.series_list)
corr_mat = np.empty((n,n))
corr_mat[:] = np.nan #let's only work on half of the matrix to speeds things up
for i in range(0, n-1): # loop over records
ts = mgs.series_list[i]
mgsl = pyleo.MultipleGeoSeries(mgs.series_list[i+1:]) #gather potential pairs
corr = mgsl.correlation(ts,method='built-in') # compute correlations
corr_mat[i,i+1:] = corr.r # save result in matrix
Looping over 15 Series in collection
100%|██████████| 15/15 [00:00<00:00, 543.36it/s]
Looping over 14 Series in collection
100%|██████████| 14/14 [00:00<00:00, 612.66it/s]
Looping over 13 Series in collection
100%|██████████| 13/13 [00:00<00:00, 2594.87it/s]
Looping over 12 Series in collection
100%|██████████| 12/12 [00:00<00:00, 3536.76it/s]
Looping over 11 Series in collection
100%|██████████| 11/11 [00:00<00:00, 3804.83it/s]
Looping over 10 Series in collection
100%|██████████| 10/10 [00:00<00:00, 2108.33it/s]
Looping over 9 Series in collection
100%|██████████| 9/9 [00:00<00:00, 1791.42it/s]
Looping over 8 Series in collection
100%|██████████| 8/8 [00:00<00:00, 501.88it/s]
Looping over 7 Series in collection
100%|██████████| 7/7 [00:00<00:00, 980.70it/s]
Looping over 6 Series in collection
100%|██████████| 6/6 [00:00<00:00, 1411.43it/s]
Looping over 5 Series in collection
100%|██████████| 5/5 [00:00<00:00, 2103.25it/s]
Looping over 4 Series in collection
100%|██████████| 4/4 [00:00<00:00, 1598.13it/s]
Looping over 3 Series in collection
100%|██████████| 3/3 [00:00<00:00, 1029.53it/s]
Looping over 2 Series in collection
100%|██████████| 2/2 [00:00<00:00, 651.24it/s]
Looping over 1 Series in collection
100%|██████████| 1/1 [00:00<00:00, 21290.88it/s]
labels = []
for item in mgs.series_list:
labels.append(item.label)
core_matrix_df = pd.DataFrame(corr_mat, index=labels, columns=labels)
plt.figure(figsize=(10, 8))
sns.heatmap(core_matrix_df, annot=True, fmt=".2f", cmap="coolwarm", center=0)
plt.title("Correlation Heatmap")
plt.tight_layout()
plt.show()
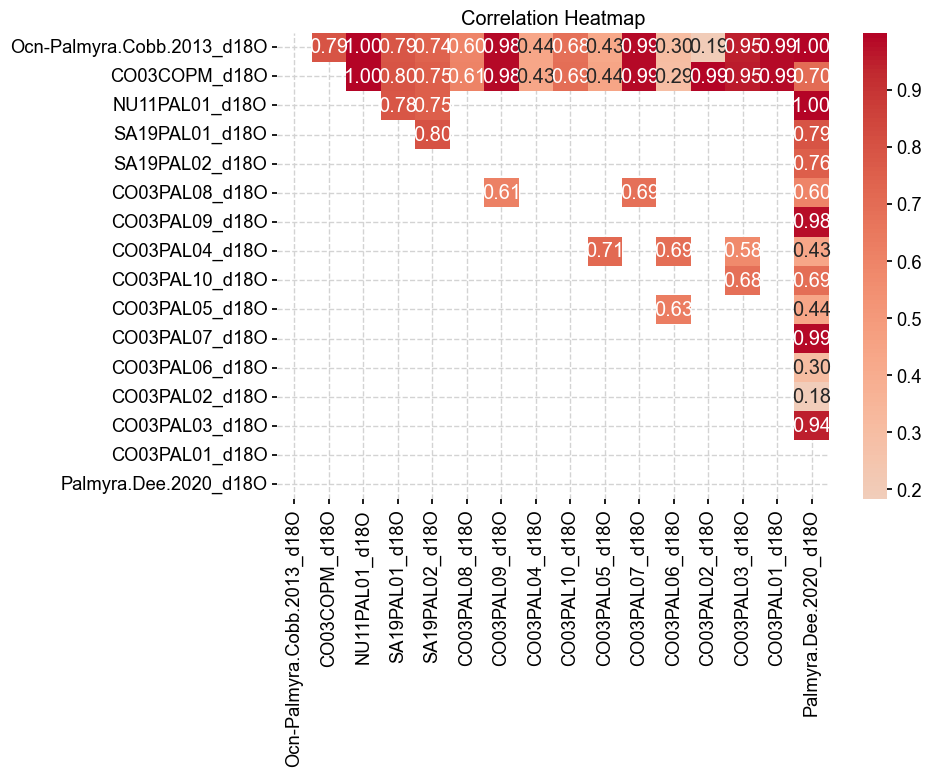
Some of the correlations are high, suggesting that the small segments went into the composited records. Note that some of the correlations were not run since the segments were not overlapping and were, therefore, skipped.
Option 2: DTW#
Let’s look at the DTW method to identify duplicates.
# --- Prepare the data ----
records = {}
times = {}
for series in mgs.series_list:
records[series.label] = series.value
times[series.label] = series.time
# --- Main loop ---
labels = list(records.keys())
n = len(labels)
dtw_matrix = np.full((n, n), np.nan)
for i, j in itertools.combinations(range(n), 2):
key_i, key_j = labels[i], labels[j]
s1, s2 = clip_to_overlap(times[key_i], records[key_i],
times[key_j], records[key_j])
if s1 is not None:
path = dtw.warping_path(s1, s2)
dist = dtw.distance(s1, s2)
norm_dist = dist / len(path) # Per-step normalization
dtw_matrix[i, j] = dtw_matrix[j, i] = norm_dist
# Convert to DataFrame for visualization
dtw_df = pd.DataFrame(dtw_matrix, index=labels, columns=labels)
plt.figure(figsize=(10, 8))
sns.heatmap(dtw_df, annot=True, fmt=".2f", cmap="viridis", mask=np.isnan(dtw_df))
plt.title("DTW Distance Matrix (Clipped to Overlap)")
plt.tight_layout()
plt.show()

The DTW method largely confirms what we have seen from the correlations in terms of duplicates. However, note that the DTW method indicates that the records from OCN-Palmyra.Cobb.2013 and CO03COPM have a correlation of 0.79 but a DTW of 0. The DTW would essentially indicate that they are in fact the same records but the correlation is less than 0.8. This is due to the interpolation needed to calculate correlation over the 14th century, highlighting the need to run DTW for records with larger gaps. We will use the insights gained here to clean the database in the next section of this notebook. For now, let’s add the Dee et al. (2020) record to the database:
df_raw = pd.concat([df_raw, df_palm_filt], ignore_index=True)
df_raw.tail()
| dataSetName | compilationName | archiveType | geo_meanLat | geo_meanLon | geo_meanElev | paleoData_variableName | paleoData_standardName | paleoData_values | paleoData_units | ... | paleoData_proxyGeneral | paleoData_seasonality | paleoData_interpName | paleoData_interpRank | TSID | time_variableName | time_standardName | time_values | time_units | distance_km | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 968 | ZI08MAY01 | CoralHydro2k | Coral | -12.6500 | 45.10 | -2.0 | SrCa | Sr/Ca | [8.947578, 8.797017, 8.784511, 8.751525, 8.778... | mmol/mol | ... | None | None | None | NaN | ZI08MAY01_SrCa | year | year | [1881.6247, 1881.791367, 1881.958033, 1882.124... | yr AD | 16936.585327 |
| 969 | LI06FIJ01 | CoralHydro2k | Coral | -16.8200 | 179.23 | -10.0 | d18O | d18O | [-4.6922, -4.6266, -4.6018, -4.5486, -4.6102, ... | permil | ... | None | None | None | NaN | LI06FIJ01_d18O | year | year | [1617.5, 1618.5, 1619.5, 1620.5, 1621.5, 1622.... | yr AD | 3250.716202 |
| 970 | SM06LKF02 | CoralHydro2k | Coral | 24.5600 | -81.41 | -4.0 | d18O | d18O | [-3.85, -3.98, -4.21, -4.06, -3.97, -4.04, -3.... | permil | ... | None | None | None | NaN | SM06LKF02_d18O | year | year | [1960.97, 1961.03, 1961.09, 1961.15, 1961.21, ... | yr AD | 8799.120973 |
| 971 | SM06LKF02 | CoralHydro2k | Coral | 24.5600 | -81.41 | -4.0 | SrCa | Sr/Ca | [9.225, nan, 9.195, 9.221, 9.198, 9.281, 9.319... | mmol/mol | ... | None | None | None | NaN | SM06LKF02_SrCa | year | year | [1960.97, 1961.03, 1961.09, 1961.15, 1961.21, ... | yr AD | 8799.120973 |
| 972 | Palmyra.Dee.2020 | None | Coral | 5.8664 | -162.12 | -9.0 | d18O | d18O | [-4.749, -4.672, -4.724, -4.717, -4.8947, -4.8... | permil | ... | isotopic | subannual | temperature | NaN | PCU-1cce3af5639a4ec | year | year | [1146.375, 1146.4583, 1146.5417, 1146.625, 114... | yr AD | NaN |
5 rows × 21 columns
Let’s drop the added columns that we put in for distance:
df_raw = df_raw.drop(columns=['distance_km'])
Data Cleaning and Pre-processing#
This section deals with cleaning and pre-processing for use with the cfr codebase. We will do so in three steps:
Step 1: Remove records that are less than 150 years. For reconstruction purposes, we need series that go back at least to 1850; many in this database do not, so we remove them to avoid disappointments later on.
Step 2: Remove duplicates. We will use an algorithm based on the data exploratory analysis we did for the specific case of the Palmyra record and generalize it.
Step 3: Filtering for records with annual to subannual resolution.
Step 1: Remove records shorter than 150 years#
For reconstruction purposes, we need series that go back at least to 1850; many in this database do not, so we remove them to avoid disappointments later on. Let’s first look at the start date of these records.
df_raw['min_time'] = df_raw['time_values'].apply(lambda x: min(x) if isinstance(x, list) and x else float('inf'))
filtered_df = df_raw[df_raw['min_time'] < 1850]
filtered_df = filtered_df.drop(columns=['min_time'])
print(filtered_df.shape)
(744, 20)
That leaves us with 744 timeseries.
Step 2: Screen for duplicates#
Based on the preliminary work we have done with the Palmyra records, we will used the following algorithm to identify duplicates:
Look for records within 10km of each other
Look at whether the variable name is the same
Calculate correlations among the records
Calculate the DTW distance among the records.
For correlations >0.8 OR DTW distance < 0.03, use the following:
a. Use the latest publication date (based on dataset name)
b. Use the longest record if the publication date is the same
Let’s have a look at the variable names present in the database to see if there are synonyms we have to deal with. Let’s make sure that we use a criteria that doesn’t have any missing values:
filtered_df.info()
<class 'pandas.core.frame.DataFrame'>
Index: 744 entries, 0 to 972
Data columns (total 20 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 dataSetName 744 non-null object
1 compilationName 743 non-null object
2 archiveType 744 non-null object
3 geo_meanLat 744 non-null float64
4 geo_meanLon 744 non-null float64
5 geo_meanElev 737 non-null object
6 paleoData_variableName 744 non-null object
7 paleoData_standardName 744 non-null object
8 paleoData_values 744 non-null object
9 paleoData_units 336 non-null object
10 paleoData_proxy 665 non-null object
11 paleoData_proxyGeneral 501 non-null object
12 paleoData_seasonality 659 non-null object
13 paleoData_interpName 665 non-null object
14 paleoData_interpRank 12 non-null float64
15 TSID 744 non-null object
16 time_variableName 744 non-null object
17 time_standardName 744 non-null object
18 time_values 744 non-null object
19 time_units 744 non-null object
dtypes: float64(3), object(17)
memory usage: 122.1+ KB
From this, it seems that paleoData_standardName could work for our purposes since there are no missing values in this field and most of the names should have been standardized:
filtered_df['paleoData_standardName'].unique()
array(['d18O', 'MXD', 'density', 'ringWidth', 'temperature',
'reflectance', 'Sr/Ca', 'iceMelt', 'd2H', 'thickness',
'RABD660670', 'calcificationRate', 'varveThickness', 'MAR'],
dtype=object)
The names are unique. Let’s proceed with the main loop. But first, let’s define some constants and parameters for the algorithm described above:
# --- Define some parameters ---
threshold = 10 # Distance in km
corr_threshold = 0.8 # Corr threshold over which two records are considered the same.
dtw_threshold = 0.03 # DTW threshold under which two records are considered the same.
Let’s also consider writing some functions to calculate the various criteria described above:
Distance on aa sphere using the Haversine distance
Correlation among multiple timeseries using Pyleoclim
DTW amon multiple timeseries
# --- Haversine distance function in km ---
def haversine(lat1, lon1, lats2, lons2, R=6371):
"""Calculate the haversine distance between a target location and a vector of locations.
Args:
lat1 (float): The target latitude
lon1 (float): the target longitude
lats2 (np.array or df.Series): The latitude of the locations to check against
lons2 (np.array or df.Series): The longitude of the locations to check against
R (int, optional): The radius of the planet in km. Defaults to 6371 for Earth.
Returns:
np.array: The distance between the target location and the vector of locations in km.
"""
# Convert degrees to radians
lat1 = np.radians(lat1)
lon1 = np.radians(lon1)
lats2 = np.radians(lats2)
lons2 = np.radians(lons2)
# Haversine formula
dlat = lats2 - lat1
dlon = lons2 - lon1
a = np.sin(dlat / 2)**2 + np.cos(lat1) * np.cos(lats2) * np.sin(dlon / 2)**2
c = 2 * np.arcsin(np.sqrt(a))
return R*c
# ----- Correlation function ------
def correlation(data):
"""Calculate correlation among multiple timeseries
Parameters
----------
data: pd.DataFrame
A pandas DataFrame containing the necessary information to create Pyleoclim.GeoSeries() objects
Returns
-------
A DataFrame containing pairwise correlations
"""
ds_name1 = [] # list of dataset names used as correlation basis
ds_name2 = [] # list of dataset names used as the series to correlate against
# Same for tsids since they are unique and we can find them again easily in the database
tsid1 = []
tsid2 = []
# list to keep the correlations
corr = []
# Create a MultipleGeoSeries object
ts_list = []
for idx, row in data.iterrows():
ts = pyleo.GeoSeries(time=row['time_values'],value=row['paleoData_values'],
time_name='Time',value_name=row['paleoData_standardName'],
time_unit=row['time_units'], value_unit=row['paleoData_units'],
lat = row['geo_meanLat'], lon = row['geo_meanLon'],
elevation = row['geo_meanElev'], archiveType = row['archiveType'],
label=row['TSID']+'+'+row['dataSetName'], verbose = False)
ts_list.append(ts)
for i,j in itertools.combinations(range(len(ts_list)), 2):
ts1 = ts_list[i] #grab the first timeseries
ts2 = ts_list[j]
# append the names and tsids
ds_name1.append(ts1.label.split('+')[1])
tsid1.append(ts1.label.split('+')[0])
ds_name2.append(ts2.label.split('+')[1])
tsid2.append(ts2.label.split('+')[0])
# calculate the correlation
corr_res = ts1.correlation(ts2, method = 'built-in')
# Store the information
corr.append(corr_res.r)
# Step 3: Summarize results
return pd.DataFrame({'dataSetName1': ds_name1,
'dataSetName2': ds_name2,
'TSID1': tsid1,
'TSID2': tsid2,
'correlation': corr})
# ------ DTW function
def dtw_pair(data):
"""Calculates the DTW betwen the records in the dataframe
Args:
data (pd.DataFrame): A pandas DataFrame containing the necessary information to create Pyleoclim.GeoSeries() objects
Returns:
pd.DataFrame: A DataFrame containing pairwise DTW
"""
### Helper function
def clip_to_overlap(time1, series1, time2, series2, min_overlap=5):
"""Helper function to clip the series to common overlap. Note we do not need to interpolate for DTW, just make sure that the time is overlapping
Args:
time1 (np.array): Time vector for Series 1
series1 (np.array): Value vector for Series 1
time2 (np.array): Time vector for Series 2
series2 (np.array): Value vector for Series 2
min_overlap (int, optional): Minimum number of pointe to even attempt the computation. Defaults to 5.
Returns:
np.array, np.array: The zscore of the values for the time of overlap.
"""
# Make sure things are sorted and have no NaNs
valid1 = (~np.isnan(time1)) & (~np.isnan(series1))
time1 = time1[valid1]
series1 = series1[valid1]
valid2 = (~np.isnan(time2)) & (~np.isnan(series2))
time2 = time2[valid2]
series2 = series2[valid2]
# Return early if either series is now empty
if len(time1) == 0 or len(time2) == 0:
return None, None
sort_idx1 = np.argsort(time1)
time1_sorted = time1[sort_idx1]
series1_sorted = series1[sort_idx1]
sort_idx2 = np.argsort(time2)
time2_sorted = time2[sort_idx2]
series2_sorted = series2[sort_idx2]
# Determine overlapping interval
start = max(time1_sorted[0], time2_sorted[0])
end = min(time1_sorted[-1], time2_sorted[-1])
# Apply mask
mask1 = (time1_sorted >= start) & (time1_sorted <= end)
mask2 = (time2_sorted >= start) & (time2_sorted <= end)
if np.sum(mask1) < min_overlap or np.sum(mask2) < min_overlap:
return None, None # Not enough overlapping points
clipped1 = series1_sorted[mask1]
clipped2 = series2_sorted[mask2]
# Avoid division by zero if std = 0
if np.std(clipped1) == 0 or np.std(clipped2) == 0:
return None, None
return zscore(clipped1), zscore(clipped2)
### Main function
ds_name1 = [] # list of dataset names used as correlation basis
ds_name2 = [] # list of dataset names used as the series to correlate against
# Same for tsids since they are unique and we can find them again easily in the database
tsid1 = []
tsid2 = []
# list to keep the DTW
dtw_list = []
for idx1, idx2 in itertools.combinations(data.index, 2):
row1 = data.loc[idx1] #grab the first record
row2 = data.loc[idx2] #grab the second record
ds_name1.append(row1['dataSetName'])
ds_name2.append(row2['dataSetName'])
tsid1.append(row1['TSID'])
tsid2.append(row2['TSID'])
s1, s2 = clip_to_overlap(np.array(row1['time_values']), np.array(row1['paleoData_values']),
np.array(row2['time_values']), np.array(row2['paleoData_values']))
if s1 is not None:
path = dtw.warping_path(s1, s2)
dist = dtw.distance(s1, s2)
dtw_list.append(dist / len(path)) # Per-step normalization
else:
dtw_list.append(np.nan)
df= pd.DataFrame({'dataSetName1': ds_name1,
'dataSetName2': ds_name2,
'TSID1': tsid1,
'TSID2': tsid2,
'dtw': dtw_list})
return df
df_list = [] # Containers for the list of DataFrames with spurious correlations
tsids = [] #to keep track of the tsids that have already been checked against the distance criteria
#make a copy of the dataframe as we need to modify it for this loop
df_copy = filtered_df.copy()
for idx, row in df_copy.iterrows():
# Check that we haven't already looked at the record in the context of another distance search
if row['TSID'] in tsids:
continue #go to the next row
# ---- Criteria 1: Distance -----
# Target location
lat1, lon1 = row['geo_meanLat'], row['geo_meanLon']
distance_km = haversine(lat1,lon1,df_copy['geo_meanLat'],df_copy['geo_meanLon'])
# Add distance column
df_copy['distance_km'] = distance_km
# Filter to only rows within a threshold
df_distance = df_copy[df_copy['distance_km'] <= threshold].reset_index(drop=True)
# Store the TSIDs so we don't run this multiple times
tsids.extend(df_distance['TSID'].to_list())
# If there is only one entry (the original one, go to the next row)
if len(df_distance)<=1:
continue
# ---- Criteria 2: Variable Name ------
var_unique = df_distance['paleoData_standardName'].unique()
for var in var_unique:
df_var_filt = df_distance[df_distance['paleoData_standardName'] == var].reset_index(drop=True)
if len(df_var_filt) >1:
# ----- Criteria 3: Correlation -------
corr_df = correlation(df_var_filt)
# ------ Criteria 4: DTW --------
dtw_df = dtw_pair(df_var_filt)
#put the results together
common_cols = ['dataSetName1', 'dataSetName1', 'TSID1','TSID2']
df_combined = pd.merge(corr_df, dtw_df, on=common_cols)
df_res = df_combined[(df_combined['correlation'] > corr_threshold) | (df_combined['dtw'] < dtw_threshold)]
if not df_res.empty:
df_list.append(df_res)
Let’s see how many similar records we have:
len(df_list)
89
Let’s have a look at each records with duplicated and create a log (human in the loop) of why records were removed from the database.
⚠️ Warning
The following cell produces a widget that allows you to go through each of the similar records and to keep/remove records. If you do so, you will change the nature of the database.
from IPython.display import display, clear_output, Markdown
import ipywidgets as widgets
# Storage
flagged_records = []
group_notes = []
df_copy = filtered_df.copy()
index_tracker = {'i': 0}
def run_iteration(i):
clear_output(wait=True)
# Final screen
if i >= len(df_list):
print("✅ Review complete!")
if flagged_records:
print("\n🚩 Flagged TSIDs:")
display(pd.DataFrame(flagged_records))
else:
print("\n🚩 No TSIDs were flagged.")
if group_notes and any(n['Note'].strip() for n in group_notes):
print("\n📝 Notes from each group:")
markdown_notes = "\n\n".join(
[
f"### Iteration {note['Group'] + 1}\n"
f"```\n{note['Note']}\n```"
for note in group_notes if note['Note'].strip()
]
)
display(Markdown(markdown_notes))
else:
print("\n📝 No group notes were entered.")
# Final Done button
done_button = widgets.Button(description="Done", button_style='success')
def on_done_clicked(b):
clear_output(wait=True)
print("🎉 Done! You may now export your results.")
done_button.on_click(on_done_clicked)
display(done_button)
return
# Begin reviewing group i
df = df_list[i]
print(f"--- Reviewing DataFrame {i+1}/{len(df_list)} ---")
valid_ids = pd.concat([df['TSID1'], df['TSID2']]).unique()
df_lookup = df_copy[df_copy['TSID'].isin(valid_ids)]
display(df_lookup)
# Plot time series
ts_list = []
for _, row in df_lookup.iterrows():
ts = pyleo.GeoSeries(
time=row['time_values'],
value=row['paleoData_values'],
time_name='Time',
value_name=row['paleoData_standardName'],
time_unit=row['time_units'],
value_unit=row['paleoData_units'],
lat=row['geo_meanLat'],
lon=row['geo_meanLon'],
elevation=row['geo_meanElev'],
archiveType=row['archiveType'],
label=row['TSID'] + '+' + row['dataSetName'],
verbose=False
)
ts_list.append(ts)
if ts_list:
mgs = pyleo.MultipleGeoSeries(ts_list)
fig, ax = mgs.stackplot()
fig.show()
else:
print("No valid time series to plot.")
# Create widgets for TSIDs
tsid_widgets = []
for tsid in valid_ids:
toggle = widgets.RadioButtons(
options=['Keep', 'Remove'],
value='Keep',
description=f'{tsid}:',
layout=widgets.Layout(width='40%')
)
tsid_widgets.append((tsid, toggle))
toggle_box = widgets.VBox([w[1] for w in tsid_widgets])
# Shared note box (pre-filled)
note_template = "Records from different samples. Not true duplicates."
note_box = widgets.Textarea(
value=note_template,
layout=widgets.Layout(width='100%', height='60px')
)
# Buttons
next_button = widgets.Button(description="Next", button_style='success')
back_button = widgets.Button(description="Back", button_style='warning')
quit_button = widgets.Button(description="Quit", button_style='danger')
button_box = widgets.HBox([next_button, back_button, quit_button])
# --- Button callbacks ---
def on_next_clicked(b):
note = note_box.value.strip()
# Remove prior flags and note for this group
group_notes[:] = [n for n in group_notes if n['Group'] != i]
flagged_records[:] = [r for r in flagged_records if r['TSID'] not in valid_ids]
# Save new notes and flags
group_notes.append({'Group': i, 'Note': note})
for tsid, toggle in tsid_widgets:
if toggle.value == 'Remove':
flagged_records.append({'TSID': tsid, 'Note': note})
index_tracker['i'] += 1
run_iteration(index_tracker['i'])
def on_back_clicked(b):
if index_tracker['i'] > 0:
index_tracker['i'] -= 1
run_iteration(index_tracker['i'])
else:
print("You're already at the first group.")
def on_quit_clicked(b):
clear_output(wait=True)
print("⚠️ Quit early.")
if flagged_records:
print("\n🚩 Flagged TSIDs:")
display(pd.DataFrame(flagged_records))
if group_notes and any(n['Note'].strip() for n in group_notes):
print("\n📝 Notes from each group:")
markdown_notes = "\n\n".join(
[
f"### Iteration {note['Group'] + 1}\n"
f"```\n{note['Note']}\n```"
for note in group_notes if note['Note'].strip()
]
)
display(Markdown(markdown_notes))
else:
print("\n📝 No group notes were entered.")
# Hook up buttons
next_button.on_click(on_next_clicked)
back_button.on_click(on_back_clicked)
quit_button.on_click(on_quit_clicked)
# Display UI
display(button_box, toggle_box, note_box)
# Start the interactive loop
run_iteration(index_tracker['i'])
| TSID | Note | |
|---|---|---|
| 0 | Ocean2kHR_142_iso2k | Ocean2kHR_142_iso2k is a duplicate of Ocean2kH... |
| 1 | Ocean2kHR_156_iso2k | Ocean2kHR_142_iso2k is a duplicate of Ocean2kH... |
| 2 | Ocean2kHR_157_iso2k | Ocean2kHR_142_iso2k is a duplicate of Ocean2kH... |
| 3 | LI00RAR01_d18O | Same records. Keeping 147. |
| 4 | LI00RAR01_SrCa | Same record. Keeping 149 |
| 5 | Ocean2kHR_006_iso2k | Same records, keeping Ocean2kHR_006 from PAGES2k |
| 6 | CH03LOM01_d18O | Same records, keeping Ocean2kHR_006 from PAGES2k |
| 7 | Ocean2kHR_019_iso2k | Same record. Keeping Ocean2kHR_019 |
| 8 | FE18RUS01_d18O | Same record. Keeping Ocean2kHR_019 |
| 9 | CA07FLI01_SrCa | Records from different samples. Not true dupli... |
| 10 | Ocean2kHR_193_iso2k | Same record. Keeping Ocean2kHR_193 |
| 11 | CA07FLI01_d18O | Same record. Keeping Ocean2kHR_193 |
| 12 | Ocean2kHR_026_iso2k | Two records but duplicates. Keeping Ocean2kHR_... |
| 13 | KU99HOU01_d18O | Two records but duplicates. Keeping Ocean2kHR_... |
| 14 | ZI14HOU01_d18O | Two records but duplicates. Keeping Ocean2kHR_... |
| 15 | ZI14HOU01_SrCa | Two records. Keeping Ocean2kHR 027 and 025 |
| 16 | Ocean2kHR_133_iso2k | Same record across all compilations. Keeping O... |
| 17 | QU96ESV01_d18O | Same record across all compilations. Keeping O... |
| 18 | Ocean2kHR_183_iso2k | Same record across all three compilations. Kee... |
| 19 | OS14UCP01_d18O | Same record across all three compilations. Kee... |
| 20 | Ocean2kHR_166_iso2k | Same record across all three compilations. Kee... |
| 21 | AS05GUA01_d18O | Same record across all three compilations. Kee... |
| 22 | KI08PAR01_d18O | Same record from two different compilations. K... |
| 23 | Ocean2kHR_173_iso2k | Same record from three different compilations.... |
| 24 | DU94URV01_d18O | Same record from three different compilations.... |
| 25 | Ocean2kHR_001_iso2k | Same record from three different compilations.... |
| 26 | CO00MAL01_d18O | Same record from three different compilations.... |
| 27 | DR99ABR01_d18O | Same record from two different compilations. K... |
| 28 | Ocean2kHR_094_iso2k | Same record from two different compilations. K... |
| 29 | Ocean2kHR_007_iso2k | Same record from three compilations (note that... |
| 30 | DA06MAF02_d18O | Same record from three compilations (note that... |
| 31 | Ocean2kHR_177_iso2k | Same record from three different compilations.... |
| 32 | UR00MAI01_d18O | Same record from three different compilations.... |
| 33 | SAm_035_iso2k | Same record from two different compilations. K... |
| 34 | Ocean2kHR_139 | Palmyra record!!!! Keeping PCU-1cce3af5639a4ec... |
| 35 | Ocean2kHR_139_iso2k | Palmyra record!!!! Keeping PCU-1cce3af5639a4ec... |
| 36 | CO03PAL08_d18O | Palmyra record!!!! Keeping PCU-1cce3af5639a4ec... |
| 37 | CO03PAL09_d18O | Palmyra record!!!! Keeping PCU-1cce3af5639a4ec... |
| 38 | CO03PAL04_d18O | Palmyra record!!!! Keeping PCU-1cce3af5639a4ec... |
| 39 | CO03PAL10_d18O | Palmyra record!!!! Keeping PCU-1cce3af5639a4ec... |
| 40 | CO03PAL05_d18O | Palmyra record!!!! Keeping PCU-1cce3af5639a4ec... |
| 41 | CO03PAL07_d18O | Palmyra record!!!! Keeping PCU-1cce3af5639a4ec... |
| 42 | CO03PAL06_d18O | Palmyra record!!!! Keeping PCU-1cce3af5639a4ec... |
| 43 | CO03PAL02_d18O | Palmyra record!!!! Keeping PCU-1cce3af5639a4ec... |
| 44 | CO03PAL03_d18O | Palmyra record!!!! Keeping PCU-1cce3af5639a4ec... |
| 45 | CO03PAL01_d18O | Palmyra record!!!! Keeping PCU-1cce3af5639a4ec... |
| 46 | Ocean2kHR_020_iso2k | Same record from different compilations. Keepi... |
| 47 | ZI04IFR01_d18O | Same record from different compilations. Keepi... |
| 48 | DE12ANC01_SrCa | Same record from two different compilations. K... |
| 49 | WE09ARR01_d18O | Same record from two different compilations. K... |
| 50 | WE09ARR01_SrCa | Same record from two different compilations. K... |
| 51 | Ocean2kHR_013_iso2k | Same record from two different compilations. K... |
| 52 | Ocean2kHR_126_iso2k | Same record from different compilations. Keepi... |
| 53 | Ocean2kHR_105_iso2k | Same reocords. Keeping Ocean2kHR 102 and 105 a... |
| 54 | Ocean2kHR_102_iso2k | Same reocords. Keeping Ocean2kHR 102 and 105 a... |
| 55 | Ocean2kHR_154_iso2k | Same record from different compilation. Keepin... |
| 56 | GO12SBV01_d18O | Same record from different compilation. Keepin... |
| 57 | WU13TON01_SrCa | Same record from different compilations. Keepi... |
| 58 | Ocean2kHR_138_iso2k | Same record from different compilations. Keepi... |
| 59 | LI94SEC01_d18O | Same record from different compilations. Keepi... |
Iteration 1
Records from different samples. Not true duplicates.
Iteration 2
Records from different samples. Not true duplicates.
Iteration 3
Records from different samples. Not true duplicates.
Iteration 4
Records from different samples. Not true duplicates.
Iteration 5
Records from different samples. Not true duplicates.
Iteration 6
Records from different samples. Not true duplicates.
Iteration 7
Records from different samples. Not true duplicates.
Iteration 8
Records from different samples. Not true duplicates.
Iteration 9
Ocean2kHR_142_iso2k is a duplicate of Ocean2kHR_142 deom most likely PAGES2k. Removing iso2k record. Same with 156 and 157
Iteration 10
From publications info in GraphDB. Different records.
Iteration 11
Same records. Keeping 147.
Iteration 12
Same record. Keeping 149
Iteration 13
Same records, keeping Ocean2kHR_006 from PAGES2k
Iteration 14
Same record. Keeping Ocean2kHR_019
Iteration 15
Records from different samples. Not true duplicates.
Iteration 16
Records from different samples. Not true duplicates.
Iteration 17
Same record. Keeping Ocean2kHR_193
Iteration 18
Two records but duplicates. Keeping Ocean2kHR_023 and 026.
Iteration 19
Two records. Keeping Ocean2kHR 027 and 025
Iteration 20
Same record across all compilations. Keeping Ocean2kHR_133
Iteration 21
Records from different samples. Not true duplicates.
Iteration 22
Same record across all three compilations. Keeping Ocean2kHR_183
Iteration 23
Same record across all three compilations. Keeping Ocean2kHR166
Iteration 24
Records from different samples. Not true duplicates.
Iteration 25
Records from different samples. Not true duplicates.
Iteration 26
Records from different samples. Not true duplicates.
Iteration 27
Records from different samples. Not true duplicates.
Iteration 28
Records from different samples. Not true duplicates.
Iteration 29
Records from different samples. Not true duplicates.
Iteration 30
Same record from two different compilations. Keeping Ocean2kHR_099
Iteration 31
Same record from two different compilations. Keeping Ocean2HR_100
Iteration 32
Records from different samples. Not true duplicates.
Iteration 33
Records from different samples. Not true duplicates.
Iteration 34
Same record from three different compilations. Keeping Ocean2kHR_173
Iteration 35
Records from different samples. Not true duplicates.
Iteration 36
Same record from three different compilations. Keeping Ocean2kHR_001
Iteration 37
Records from different samples. Not true duplicates.
Iteration 38
Records from different samples. Not true duplicates.
Iteration 39
Records from different samples. Not true duplicates.
Iteration 40
Same record from two different compilations. Keeping Ocean2kHR_167
Iteration 41
Same record from two different compilations. Keeping Ocean2kHR_094
Iteration 42
Same record from three compilations (note that it's most likely from two different coral head which CoralHydro2k archived in different dataset). Keeping Ocean2kHR_007
Iteration 43
Records from different samples. Not true duplicates.
Iteration 44
Records from different samples. Not true duplicates.
Iteration 45
Records from different samples. Not true duplicates.
Iteration 46
Records from different samples. Not true duplicates.
Iteration 47
Same record from three different compilations. Keeping Ocean2kHR_177
Iteration 48
Records from different samples. Not true duplicates.
Iteration 49
Same record from two different compilations. Keeping SAm_035
Iteration 50
Records from different samples. Not true duplicates.
Iteration 51
Records from different samples. Not true duplicates.
Iteration 52
Records from different samples. Not true duplicates.
Iteration 53
Palmyra record!!!! Keeping PCU-1cce3af5639a4ec from Dee et al. (2020)
Iteration 54
Records from different samples. Not true duplicates.
Iteration 55
Records from different samples. Not true duplicates.
Iteration 56
Records from different samples. Not true duplicates.
Iteration 57
Records from different samples. Not true duplicates.
Iteration 58
Records from different samples. Not true duplicates.
Iteration 59
Records from different samples. Not true duplicates.
Iteration 60
Records from different samples. Not true duplicates.
Iteration 61
Records from different samples. Not true duplicates.
Iteration 62
Same record from different compilations. Keeping Ocean2kHR_020
Iteration 63
Records from different samples. Not true duplicates.
Iteration 64
Records from different samples. Not true duplicates.
Iteration 65
Records from different samples. Not true duplicates.
Iteration 66
Same record from two different compilations. Keeping Ocean2kHR_134
Iteration 67
Records from different samples. Not true duplicates.
Iteration 68
Records from different samples. Not true duplicates.
Iteration 69
Records from different samples. Not true duplicates.
Iteration 70
Records from different samples. Not true duplicates.
Iteration 71
Same record from two different compilations. Keeping Ocean2kHR_190
Iteration 72
Same record from two different compilations. Keeping Ocean2kHR_191
Iteration 73
Records from different samples. Not true duplicates.
Iteration 74
Records from different samples. Not true duplicates.
Iteration 75
Same record from two different compilations. Keeping Ocean2kHR_013
Iteration 76
Records from different samples. Not true duplicates.
Iteration 77
Records from different samples. Not true duplicates.
Iteration 78
Records from different samples. Not true duplicates.
Iteration 79
Records from different samples. Not true duplicates.
Iteration 80
Same record from different compilations. Keeping Ocean2kHR_126
Iteration 81
Same reocords. Keeping Ocean2kHR 102 and 105 as well as DR00NBB01, which seems to be unique
Iteration 82
Records from different samples. Not true duplicates.
Iteration 83
Same record from different compilation. Keeping Ocean2kHR_154
Iteration 84
Same record from different compilations. Keeping Ocean2kHR_159
Iteration 85
Records from different samples. Not true duplicates.
Iteration 86
Same record from different compilations. Keeping Ocean2kHR_138
Iteration 87
Records from different samples. Not true duplicates.
Iteration 88
Records from different samples. Not true duplicates.
Iteration 89
Records from different samples. Not true duplicates.
# Save flagged TSIDs and notes
df_flags = pd.DataFrame(flagged_records)
df_flags.to_csv("./data/flagged_tsids_with_notes.csv", index=False)
df_notes = pd.DataFrame(group_notes)
df_notes.to_csv("./data/group_notes.csv", index=False)
# Cleaned dataset
df_cleaned = df_copy[~df_copy['TSID'].isin(df_flags['TSID'])]
# save the file
df_cleaned.to_pickle("./data/cleaned_timeseries.pkl")
Building the cfr.ProxyDataBase#
Reopen the file:
df_cleaned = pd.read_pickle("./data/cleaned_timeseries.pkl")
df_cleaned.head()
| dataSetName | compilationName | archiveType | geo_meanLat | geo_meanLon | geo_meanElev | paleoData_variableName | paleoData_standardName | paleoData_values | paleoData_units | paleoData_proxy | paleoData_proxyGeneral | paleoData_seasonality | paleoData_interpName | paleoData_interpRank | TSID | time_variableName | time_standardName | time_values | time_units | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Ant-WDC05A.Steig.2013 | Pages2kTemperature | Glacier ice | -79.46 | -112.09 | 1806.0 | d18O | d18O | [-33.32873325, -35.6732, -33.1574, -34.2854, -... | permil | d18O | None | Annual | temperature | NaN | Ant_030 | year | year | [2005, 2004, 2003, 2002, 2001, 2000, 1999, 199... | yr AD |
| 1 | NAm-MtLemon.Briffa.2002 | Pages2kTemperature | Wood | 32.50 | -110.80 | 2700.0 | MXD | MXD | [0.968, 0.962, 1.013, 0.95, 1.008, 0.952, 1.02... | None | maximum latewood density | dendrophysical | Summer | temperature | NaN | NAm_568 | year | year | [1568, 1569, 1570, 1571, 1572, 1573, 1574, 157... | yr AD |
| 2 | Arc-Arjeplog.Bjorklund.2014 | Pages2kTemperature | Wood | 66.30 | 18.20 | 800.0 | density | density | [-0.829089212152348, -0.733882889924006, -0.89... | None | maximum latewood density | dendrophysical | Jun-Aug | temperature | NaN | Arc_060 | year | year | [1200, 1201, 1202, 1203, 1204, 1205, 1206, 120... | yr AD |
| 3 | Asi-CHIN019.Li.2010 | Pages2kTemperature | Wood | 29.15 | 99.93 | 2150.0 | ringWidth | ringWidth | [1.465, 1.327, 1.202, 0.757, 1.094, 1.006, 1.2... | None | ring width | dendrophysical | Annual | temperature | NaN | Asia_041 | year | year | [1509, 1510, 1511, 1512, 1513, 1514, 1515, 151... | yr AD |
| 4 | NAm-Landslide.Luckman.2006 | Pages2kTemperature | Wood | 60.20 | -138.50 | 800.0 | ringWidth | ringWidth | [1.123, 0.841, 0.863, 1.209, 1.139, 1.056, 0.8... | None | ring width | dendrophysical | Summer | temperature | NaN | NAm_1876 | year | year | [913, 914, 915, 916, 917, 918, 919, 920, 921, ... | yr AD |
Adding ptype and seasonality info#
LiPD files contain seasonality (in months or phrases) and the necessary information to create a cfr ‘ptype’. The ‘ptype’ is simply the proxy archive and variable name, for example d18O of coral would be coral.d18O. The seasonality on the other hand, requires some conversion between month to the numerical list format that cfr accepts. For example, for a growing season of June, July, August, the cfr seasonality will be [6,7,8]. In the following two functions convert_seasonality and create_ptype, we can get the original dataFrame columns into the format we need for assimilation.
def convert_seasonality(seasonality_str, latitude = None):
"""
Convert LiPD seasonality strings to CFR month lists
CFR expects lists like [6,7,8] for JJA, [12,1,2] for DJF, etc.
Args:
seasonality_str: String representation of seasonality from LiPD
Returns:
list: List of month integers, or list(range(1,13)) for annual/None
"""
# Default to annual for None/NaN
if pd.isna(seasonality_str) or seasonality_str == '' or seasonality_str is None:
return list(range(1, 13))
seasonality_str = str(seasonality_str).strip()
# Hemisphere-aware mapping for Summer/Winter and Growing season
if seasonality_str in ['Summer', 'Winter'] and latitude is not None:
if latitude > 0: # NH
if seasonality_str == 'Summer':
return [6, 7, 8] # JJA
elif seasonality_str == 'Winter':
return [12, 1, 2] # DJF
elif latitude < 0: # SH
if seasonality_str == 'Summer':
return [12, 1, 2] # DJF in NH = Summer in SH
elif seasonality_str == 'Winter':
return [6, 7, 8] # JJA in NH = Winter in SH
if seasonality_str in ['Growing Season'] and latitude is not None:
if latitude > 0: # NH
return [4, 5, 6, 7, 8, 9] # Apr-Sept
else: # SH
return [10, 11, 12, 1, 2, 3] # Oct-Mar
# Exact mapping for your specific seasonality patterns
seasonality_map = {
'Annual': list(range(1, 13)),
'Jun-Aug': [6, 7, 8],
'Nov-Feb': [11, 12, 1, 2],
'May-Apr': [5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4], # May to next April
'Jun-Jul': [6, 7],
'Mar-May': [3, 4, 5],
'subannual': list(range(1, 13)), # Default to annual??
'Sep-Apr': [9, 10, 11, 12, 1, 2, 3, 4],
'Jul': [7],
'Dec-Feb': [12, 1, 2],
'Apr': [4],
'Jul-Jun': [7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6], # Jul to next June
'Sep-Nov': [9, 10, 11],
'Nov-Oct': [11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10], # Nov to next Oct
'Apr-Jul': [4, 5, 6, 7],
'Jul-Sep': [7, 8, 9],
'Oct-Apr': [10, 11, 12, 1, 2, 3, 4],
'Nov-Apr': [11, 12, 1, 2, 3, 4],
'Apr-Sep': [4, 5, 6, 7, 8, 9],
'Dec-Mar': [12, 1, 2, 3],
'Aug-Jul': [8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7], # Aug to next July
'Jun-Sep': [6, 7, 8, 9],
'Sep-Aug': [9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8], # Sep to next Aug
'Mar-Aug': [3, 4, 5, 6, 7, 8],
'Jun': [6],
'May-Sep': [5, 6, 7, 8, 9],
'Feb-Aug': [2, 3, 4, 5, 6, 7, 8],
'Jul-Dec': [7, 8, 9, 10, 11, 12],
'Mar-Oct': [3, 4, 5, 6, 7, 8, 9, 10],
'May-Jul': [5, 6, 7],
'Jan-Mar': [1, 2, 3],
'Apr-Jun': [4, 5, 6],
}
# Direct lookup
if seasonality_str in seasonality_map:
return seasonality_map[seasonality_str]
# If no pattern matches, default to annual
print(f"Warning: Could not parse seasonality '{seasonality_str}', defaulting to annual")
return list(range(1, 13))
def create_ptype(archive_type, proxy_standard_name):
"""
Determine CFR proxy type from LiPD archive and proxy information
Args:
archive_type: LiPD archiveType
proxy_standard_name: LiPD paleoData_standardName
Returns:
str: CFR-compatible proxy type string
"""
archive = str(archive_type).lower() if pd.notna(archive_type) else 'unknown'
proxy = str(proxy_standard_name).lower() if pd.notna(proxy_standard_name) else 'unknown'
if 'wood' in archive or 'tree' in archive:
if 'ring width' in proxy or 'trw' in proxy:
return 'tree.TRW'
elif 'density' in proxy or 'mxd' in proxy:
return 'tree.MXD'
else:
return 'tree.TRW'
elif 'coral' in archive:
if 'd18o' in proxy:
return 'coral.d18O'
elif 'sr/ca' in proxy:
return 'coral.SrCa'
elif 'calcification' in proxy:
return 'coral.calc'
else:
return 'coral.d18O'
elif 'ice' in archive:
if 'dD' in proxy or 'd2H' in proxy:
return 'ice.dD'
elif 'melt' in proxy:
return 'ice.melt'
else:
return 'ice.d18O'
elif 'lake' in archive:
if 'varve thickness' in proxy:
return 'lake.varve_thickness'
elif 'varve' in proxy and 'thickness' not in proxy:
return 'lake.varve_property'
elif 'chironomid' in proxy:
return 'lake.chironomid'
elif 'midge' in proxy:
return 'lake.midge'
elif 'reflectance' in proxy:
return 'lake.reflectance'
elif 'bsi' in proxy:
return 'lake.BSi'
elif 'accumulation' in proxy:
return 'lake.accumulation'
else:
return 'lake.other'
elif 'marine' in archive:
if 'alkenone' in proxy:
return 'marine.alkenone'
elif 'mg/ca' in proxy:
return 'marine.MgCa'
else:
return 'marine.other'
elif 'documents' in archive:
return 'documents'
elif 'borehole' in archive:
return 'borehole'
elif 'speleothem' in archive:
return 'speleothem.d18O'
elif 'sclerosponge' in archive:
return 'sclerosponge.d18O'
elif 'bivalve' in archive:
return 'bivalve.d18O'
else:
return 'hybrid'
Paleodata can sometimes be published in descending time order rather than ascending. This creates a problem for us since the cfr.ProxyRecord.dt or the median time difference between data points will register the dt < 0 when the time values are in descending order. To prevent this issue before creating any proxy databases, we will use flip_df to reverse the time values that are incorrect to keep it consistent.
def flip_df(df):
"""
Fix reverse time series in DataFrame
"""
df_fixed = df.copy()
fixed_count = 0
for idx, row in df_fixed.iterrows():
time_values = np.array(row['time_values'])
paleo_values = np.array(row['paleoData_values'])
if len(time_values) > 1:
# Check if time is descending (reverse)
if time_values[0] > time_values[-1]:
# Flip both arrays
df_fixed.at[idx, 'time_values'] = time_values[::-1]
df_fixed.at[idx, 'paleoData_values'] = paleo_values[::-1]
fixed_count += 1
print(f"Fixed {fixed_count} reverse time series in DataFrame")
return df_fixed
# apply the seasonality into new column
df_cleaned['cfr_seasonality'] = df_cleaned.apply(lambda row: convert_seasonality(row['paleoData_seasonality'], row['geo_meanLat']),
axis=1
)
# apply ptype conversion into new column
df_cleaned['cfr_ptype'] = df_cleaned.apply(
lambda row: create_ptype(row['archiveType'], row['paleoData_standardName']),
axis=1
)
# Create new dataframe with correctly reversed time/data values
df_cleaned_flip = flip_df(df_cleaned)
Fixed 173 reverse time series in DataFrame
cfr.ProxyDatabase object takes in very specific parameters such as latitude, longitude, elevation, ptype, seasonality, and of course the time and data value. All of this is provided in the dataFrame, so create_pdb will seamlessly convert our necessary df columns into a cfr.Proxydatabase with all necessary data to be able to work in a data assimilation workflow.
def create_pdb(df):
"""
Create CFR ProxyDatabase from DataFrame that already has cfr_seasonality and cfr_ptype columns
Args:
df: DataFrame with cfr_seasonality and cfr_ptype columns
Returns:
cfr.ProxyDatabase: Ready-to-use CFR ProxyDatabase with seasonality
"""
print("Creating CFR ProxyDatabase from enhanced DataFrame...")
records_list = []
skipped_records = 0
for idx, row in df.iterrows():
if idx % 100 == 0:
print(f" Processing {idx}/{len(df)}...")
try:
# Get the complete time and value arrays directly
time_values = np.array(row['time_values'])
paleo_values = np.array(row['paleoData_values'])
# Basic validation to make sure lengths match
if len(time_values) != len(paleo_values) or len(time_values) == 0:
skipped_records += 1
continue
# Remove any NaN values
valid_mask = ~(np.isnan(time_values) | np.isnan(paleo_values))
time_values = time_values[valid_mask]
paleo_values = paleo_values[valid_mask]
if len(time_values) == 0:
skipped_records += 1
continue
# Basic info (LiPD's tsid = cfr's pid)
dataset_name = str(row['dataSetName']) if pd.notna(row['dataSetName']) else f"Dataset_{idx}"
tsid = str(row['TSID']) if pd.notna(row['TSID']) else f"TS_{idx}"
# Location
lat = float(row['geo_meanLat']) if pd.notna(row['geo_meanLat']) else 0.0
lon = float(row['geo_meanLon']) if pd.notna(row['geo_meanLon']) else 0.0
# Handle elevation
elev = 0.0
if pd.notna(row['geo_meanElev']):
try:
elev_val = row['geo_meanElev']
if str(elev_val).lower() not in ['na', 'nan', 'none', '']:
elev = float(elev_val)
except (ValueError, TypeError):
elev = 0.0
# Use the pre-computed CFR columns
seasonality = row['cfr_seasonality']
ptype = row['cfr_ptype']
# Optional metadata
value_name = str(row['paleoData_variableName']) if pd.notna(row['paleoData_variableName']) else None
value_unit = str(row['paleoData_units']) if pd.notna(row['paleoData_units']) else None
# Create ProxyRecord directly
record = cfr.ProxyRecord(
pid=tsid,
time=time_values,
value=paleo_values,
lat=lat,
lon=lon,
elev=elev,
ptype=ptype,
seasonality=seasonality,
value_name=value_name,
value_unit=value_unit
)
records_list.append(record)
except Exception as e:
print(f" Error processing record {idx}: {e}")
skipped_records += 1
continue
# Create empty ProxyDatabase
proxy_db = cfr.ProxyDatabase()
for record in records_list:
proxy_db += record
print(f"Created CFR ProxyDatabase with {proxy_db.nrec} records")
if skipped_records > 0:
print(f" Skipped {skipped_records} records due to data issues")
return proxy_db
Once our functions are created to go from a dataFrame into a ProxyDatabase, we will branch off into two options for the data assimilation step.
Option 1: Filter without interpolation
Create cfr.ProxyDatabase -> filter by dt <= 1 (annual + subannual only) -> save as ‘pdb_w_seasonality_annual.pkl’
Option 2: Filter with interpolation
For records with dt <= 5, interpolate to annual then refilter.
Create cfr.ProxyDatabase -> pyleo.interp -> filter by dt <=1 -> save as ‘pdb_wseasonality_interp.pkl’
Quick debug before saving ProxyDatabases#
The main thing we want to make sure here is that seasonality is preserved when we convert from a pandas Dataframe to cfr.ProxyDatabase and then to a pickle file. Here we run a quick check just to make sure the important information is saved for all the records.
print(f"\n" + "="*50)
print(" >> Converting to CFR ProxyDatabase...")
try:
unfiltered_pdb = create_pdb(df_cleaned_flip)
print(f"SUCCESS: ProxyDatabase created with {unfiltered_pdb.nrec} records")
print("Proxy types:", unfiltered_pdb.type_dict)
# Verify seasonality is preserved
print(f"\nSeasonality verification:")
seasonality_preserved = 0
for pid, record in unfiltered_pdb.records.items():
if record.seasonality is not None:
seasonality_preserved += 1
print(f"Records with seasonality: {seasonality_preserved}/{unfiltered_pdb.nrec}")
# Show a sample record
sample_pid = list(unfiltered_pdb.records.keys())[0]
sample_record = unfiltered_pdb.records[sample_pid]
print(f"\nSample CFR ProxyRecord:")
print(f" PID: {sample_record.pid}")
print(f" Type: {sample_record.ptype}")
print(f" Seasonality: {sample_record.seasonality}")
print(f" Time points: {len(sample_record.time)}")
print(f" Location: ({sample_record.lat:.2f}, {sample_record.lon:.2f})")
except Exception as e:
print(f"ERROR creating CFR ProxyDatabase: {e}")
import traceback
traceback.print_exc()
==================================================
>> Converting to CFR ProxyDatabase...
Creating CFR ProxyDatabase from enhanced DataFrame...
Processing 0/684...
Processing 100/684...
Processing 200/684...
Processing 300/684...
Processing 400/684...
Processing 500/684...
Processing 600/684...
Created CFR ProxyDatabase with 684 records
SUCCESS: ProxyDatabase created with 684 records
Proxy types: {'ice.d18O': 47, 'tree.MXD': 59, 'tree.TRW': 355, 'lake.other': 42, 'marine.other': 57, 'documents': 15, 'lake.reflectance': 1, 'coral.SrCa': 33, 'coral.d18O': 58, 'ice.melt': 2, 'borehole': 3, 'speleothem.d18O': 4, 'coral.calc': 2, 'sclerosponge.d18O': 5, 'hybrid': 1}
Seasonality verification:
Records with seasonality: 684/684
Sample CFR ProxyRecord:
PID: Ant_030
Type: ice.d18O
Seasonality: [1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12]
Time points: 1220
Location: (-79.46, 247.91)
Option 1: Filter by dt <= 1#
This option is more direct, as we will use cfr’s capabilities alone to restrict the Proxy Database to only annual and subannual records. Then we will save it as a pickle file as to preserve the seasonality and also as a netCDF (for future purposes).
# Option 1 with filtering by resolution (annual and subannual only)
filtered_pdb_1 = unfiltered_pdb.filter(by='dt', keys=(0.0,1.2))
len(filtered_pdb_1.records)
559
# Save as NetCDF (without seasonality preserved)
filtered_pdb_1.to_nc('./prev_data/pdb_w_seasonality_annual.nc')
>>> Warning: this is an experimental feature.
>>> Warning: this is an experimental feature.
100%|██████████| 559/559 [00:20<00:00, 27.57it/s]
>>> Data before 1 CE is dropped for records: ['Arc_032', 'SAm_025', 'Arc_025', 'Arc_002', 'Aus_041', 'Arc_031', 'Eur_017', 'Arc_021', 'NAm_880', 'Ant_031dD', 'Ant_031', 'NAm_664', 'Ant_022', 'Arc_020', 'Eur_014', 'Arc_004'].
ProxyDatabase saved to: ./prev_data/pdb_w_seasonality_annual.nc
pickle_path = './prev_data/pdb_w_seasonality_annual.pkl'
with open(pickle_path, 'wb') as f:
pickle.dump(filtered_pdb_1, f)
# Use pickle to save proxyDatabase
print("Saving with seasonality preservation using pickle...")
print(f"✓ Saved to pickle: {pickle_path}")
# Test loading from pickle
with open(pickle_path, 'rb') as f:
test_db_pickle = pickle.load(f)
print(f"✓ Loaded from pickle: {test_db_pickle.nrec} records")
# Verify seasonality is preserved in pickle
seasonality_pickle = {}
for pid, record in test_db_pickle.records.items():
if record.seasonality is None:
key = "None"
elif record.seasonality == list(range(1, 13)):
key = "Annual [1-12]"
elif record.seasonality == [6, 7, 8]:
key = "Summer [6, 7, 8]"
elif record.seasonality == [12, 1, 2]:
key = "Winter [12, 1, 2]"
else:
key = str(record.seasonality)
seasonality_pickle[key] = seasonality_pickle.get(key, 0) + 1
print(f"\nSeasonality distribution in pickle-loaded database:")
for season, count in sorted(seasonality_pickle.items(), key=lambda x: x[1], reverse=True)[:10]:
print(f" {season}: {count}")
# Show examples
print(f"\nExample records from pickle with their seasonality:")
count = 0
for pid, record in test_db_pickle.records.items():
if count >= 10:
break
if record.seasonality != list(range(1, 13)) and record.seasonality is not None:
print(f" {record.pid}: {record.ptype} -> {record.seasonality}")
count += 1
# Verify counts
pickle_custom_seasonality = sum(1 for r in test_db_pickle.records.values()
if r.seasonality is not None and r.seasonality != list(range(1, 13)))
pickle_none_count = sum(1 for r in test_db_pickle.records.values() if r.seasonality is None)
# Debugging checks
print(f"\nPickle verification:")
print(f" Records with custom seasonality: {pickle_custom_seasonality}")
print(f" Records with None seasonality: {pickle_none_count}")
print(f" Records with annual seasonality: {test_db_pickle.nrec - pickle_custom_seasonality - pickle_none_count}")
if pickle_custom_seasonality > 0:
print(f"SUCCESS: Seasonality preserved in pickle format!")
else:
print(f"\n⚠️ Still having issues with seasonality preservation")
print(f"\n" + "="*60)
print(f"File: {pickle_path}")
Saving with seasonality preservation using pickle...
✓ Saved to pickle: ./prev_data/pdb_w_seasonality_annual.pkl
✓ Loaded from pickle: 559 records
Seasonality distribution in pickle-loaded database:
Summer [6, 7, 8]: 307
Annual [1-12]: 213
[6, 7]: 6
[5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4]: 4
[9, 10, 11, 12, 1, 2, 3, 4]: 4
Winter [12, 1, 2]: 4
[7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6]: 2
[7, 8, 9]: 2
[6]: 2
[7]: 2
Example records from pickle with their seasonality:
NAm_568: tree.MXD -> [6, 7, 8]
Arc_060: tree.MXD -> [6, 7, 8]
NAm_1876: tree.TRW -> [6, 7, 8]
NAm_1108: tree.TRW -> [6, 7, 8]
NAm_1120: tree.MXD -> [6, 7, 8]
Asia_102: tree.TRW -> [6, 7, 8]
SAm_025: lake.other -> [11, 12, 1, 2]
NAm_1396: tree.TRW -> [6, 7, 8]
NAm_220: tree.TRW -> [6, 7, 8]
SAm_020: tree.TRW -> [5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4]
Pickle verification:
Records with custom seasonality: 346
Records with None seasonality: 0
Records with annual seasonality: 213
✅ SUCCESS: Seasonality preserved in pickle format!
Use pickle format for now until CFR NetCDF seasonality support is fixed.
============================================================
File: ./prev_data/pdb_w_seasonality_annual.pkl
Option 2: Interpolated then filtered by annual/subannual#
This step is more involved than the last. Firstly we will take all records within a median of 5 year resolution. Then we will use Pyleoclim’s pyleo.interp to interpolate those resolutions to make them annual. Then in order to preserve their original time and data values, we will duplicate only the rows (matching ‘TSID’) of those records with dt<5 such that we have one that is annual and one that is the original. Once that is done, we can re implement the annual/subannual filter. For each interpolated record, the code will output the new number of data points to confirm that it does what is required.
to_interp = unfiltered_pdb.filter(by='dt', keys=(1.1, 5.1))
We take record of the TSIDs of the records that are being interpolated so we can duplicate and preserve their information later in an unaltered format.
# Create a list to store new interpolated rows
new_rows = []
failed_reasons = []
for pobj in to_interp.records:
if pobj in df_cleaned_flip['TSID'].values:
# Find the matching row
row_idx = df_cleaned_flip[df_cleaned_flip['TSID'] == pobj].index[0]
original_row = df_cleaned_flip.loc[row_idx].copy()
# Get the data
x = np.array(original_row['time_values'])
y = np.array(original_row['paleoData_values'])
print(f"\nTrying {pobj}:")
print(f" Original data: {len(x)} points, range {x.min():.1f} to {x.max():.1f}")
# Clean data (remove NaNs)
valid_mask = ~(np.isnan(x) | np.isnan(y))
x_clean = x[valid_mask]
y_clean = y[valid_mask]
print(f" After cleaning: {len(x_clean)} points")
if len(x_clean) < 3:
failed_reasons.append(f"{pobj}: Not enough points ({len(x_clean)})")
continue
# Check if data is sorted
if not np.all(np.diff(x_clean) > 0):
# Sort the data
sort_idx = np.argsort(x_clean)
x_clean = x_clean[sort_idx]
y_clean = y_clean[sort_idx]
print(f" Data was unsorted, now sorted")
try:
# Calculate step manually to avoid pyleoclim's step calculation
time_diffs = np.diff(x_clean)
median_step = np.median(time_diffs)
target_step = 1.0 # Annual
print(f" Current median step: {median_step:.3f}, target: {target_step}")
# Use explicit start/stop/step
start_time = np.ceil(x_clean.min())
stop_time = np.floor(x_clean.max())
print(f" Interpolation range: {start_time} to {stop_time}")
if stop_time <= start_time:
failed_reasons.append(f"{pobj}: Invalid time range")
continue
# Try pyleoclim with explicit parameters
x_interp, y_interp = pyleo.utils.tsutils.interp(
x_clean, y_clean,
interp_type='linear',
start=start_time,
stop=stop_time,
step=target_step,
bounds_error=False,
fill_value='extrapolate'
)
print(f" SUCCESS: {len(x_interp)} interpolated points")
# Create duplicate with interpolated data
interp_row = original_row.copy()
interp_row['time_values'] = x_interp
interp_row['paleoData_values'] = y_interp
interp_row['TSID'] = str(original_row['TSID']) + '_interp'
new_rows.append(interp_row)
except Exception as e:
failed_reasons.append(f"{pobj}: {str(e)}")
print(f" FAILED: {e}")
continue
print(f"\n=== RESULTS ===")
print(f"Successfully interpolated: {len(new_rows)} records")
print(f"Failed: {len(failed_reasons)} records")
for reason in failed_reasons[:5]: # Show first 5 failures
print(f" {reason}")
Trying O2kLR_045:
Original data: 516 points, range 1.5 to 2001.0
After cleaning: 516 points
Current median step: 4.000, target: 1.0
Interpolation range: 2.0 to 2001.0
SUCCESS: 2000 interpolated points
Trying Ocean2kHR_194:
Original data: 57 points, range 1708.0 to 1988.0
After cleaning: 57 points
Current median step: 5.000, target: 1.0
Interpolation range: 1708.0 to 1988.0
SUCCESS: 281 interpolated points
Trying Ocean2kHR_193:
Original data: 57 points, range 1708.0 to 1988.0
After cleaning: 57 points
Current median step: 5.000, target: 1.0
Interpolation range: 1708.0 to 1988.0
SUCCESS: 281 interpolated points
Trying Eur_001:
Original data: 717 points, range -90.0 to 1935.0
After cleaning: 717 points
Data was unsorted, now sorted
Current median step: 2.500, target: 1.0
Interpolation range: -90.0 to 1935.0
SUCCESS: 2026 interpolated points
Trying Ant_009:
Original data: 231 points, range 423.8 to 1467.2
After cleaning: 231 points
Current median step: 4.440, target: 1.0
Interpolation range: 424.0 to 1467.0
SUCCESS: 1044 interpolated points
Trying NAm_3870:
Original data: 2678 points, range -6015.0 to 1716.0
After cleaning: 2678 points
Data was unsorted, now sorted
Current median step: 2.000, target: 1.0
Interpolation range: -6015.0 to 1716.0
SUCCESS: 7732 interpolated points
Trying LPD335d863f:
Original data: 404 points, range 1140.4 to 2001.3
After cleaning: 404 points
Current median step: 1.871, target: 1.0
Interpolation range: 1141.0 to 2001.0
SUCCESS: 861 interpolated points
Trying O2kLR_150:
Original data: 82 points, range 1568.0 to 1983.0
After cleaning: 82 points
Current median step: 5.000, target: 1.0
Interpolation range: 1568.0 to 1983.0
SUCCESS: 416 interpolated points
Trying Ocean2kHR_125:
Original data: 25 points, range 1833.0 to 1904.0
After cleaning: 25 points
Current median step: 3.000, target: 1.0
Interpolation range: 1833.0 to 1904.0
SUCCESS: 72 interpolated points
Trying LPDb7ba9166:
Original data: 165 points, range 74.0 to 2001.0
After cleaning: 165 points
Data was unsorted, now sorted
Current median step: 3.000, target: 1.0
Interpolation range: 74.0 to 2001.0
SUCCESS: 1928 interpolated points
Trying Arc_056:
Original data: 397 points, range 3.0 to 1983.0
After cleaning: 397 points
Current median step: 5.000, target: 1.0
Interpolation range: 3.0 to 1983.0
SUCCESS: 1981 interpolated points
Trying Ocean2kHR_129:
Original data: 134 points, range 1384.0 to 1992.0
After cleaning: 113 points
Current median step: 5.000, target: 1.0
Interpolation range: 1384.0 to 1992.0
SUCCESS: 609 interpolated points
Trying Ocean2kHR_128:
Original data: 133 points, range 1389.0 to 1992.0
After cleaning: 118 points
Current median step: 5.000, target: 1.0
Interpolation range: 1389.0 to 1992.0
SUCCESS: 604 interpolated points
Trying Eur_008:
Original data: 875 points, range -1949.0 to 2000.0
After cleaning: 875 points
Data was unsorted, now sorted
Current median step: 3.000, target: 1.0
Interpolation range: -1949.0 to 2000.0
SUCCESS: 3950 interpolated points
Trying NAm_3867:
Original data: 197 points, range 1120.0 to 1900.0
After cleaning: 197 points
Current median step: 4.000, target: 1.0
Interpolation range: 1120.0 to 1900.0
SUCCESS: 781 interpolated points
Trying NAm_3868:
Original data: 197 points, range 1120.0 to 1900.0
After cleaning: 197 points
Current median step: 4.000, target: 1.0
Interpolation range: 1120.0 to 1900.0
SUCCESS: 781 interpolated points
Trying Ocean2kHR_126:
Original data: 187 points, range 1344.0 to 1991.0
After cleaning: 175 points
Current median step: 3.000, target: 1.0
Interpolation range: 1344.0 to 1991.0
SUCCESS: 648 interpolated points
Trying Ocean2kHR_127:
Original data: 184 points, range 1356.0 to 1991.0
After cleaning: 170 points
Current median step: 3.000, target: 1.0
Interpolation range: 1356.0 to 1991.0
SUCCESS: 636 interpolated points
Trying Arc_043:
Original data: 395 points, range 1.0 to 1971.0
After cleaning: 395 points
Current median step: 5.000, target: 1.0
Interpolation range: 1.0 to 1971.0
SUCCESS: 1971 interpolated points
Trying LPDe51d17e8:
Original data: 566 points, range 1221.0 to 1990.0
After cleaning: 566 points
Current median step: 1.480, target: 1.0
Interpolation range: 1222.0 to 1990.0
SUCCESS: 769 interpolated points
Trying Ant_035:
Original data: 1160 points, range -993.4 to 1937.1
After cleaning: 1055 points
Current median step: 2.500, target: 1.0
Interpolation range: -993.0 to 1937.0
SUCCESS: 2931 interpolated points
Trying Ocean2kHR_125_iso2k:
Original data: 25 points, range 1833.0 to 1904.0
After cleaning: 25 points
Current median step: 3.000, target: 1.0
Interpolation range: 1833.0 to 1904.0
SUCCESS: 72 interpolated points
Trying Ocean2kHR_128_iso2k:
Original data: 133 points, range 1389.0 to 1992.0
After cleaning: 118 points
Current median step: 5.000, target: 1.0
Interpolation range: 1389.0 to 1992.0
SUCCESS: 604 interpolated points
Trying DR00NBB01_d18O:
Original data: 25 points, range 1833.0 to 1904.0
After cleaning: 25 points
Current median step: 3.000, target: 1.0
Interpolation range: 1833.0 to 1904.0
SUCCESS: 72 interpolated points
Trying RO19PAR01_SrCa:
Original data: 97 points, range 1445.5 to 1669.5
After cleaning: 97 points
Current median step: 1.500, target: 1.0
Interpolation range: 1446.0 to 1669.0
SUCCESS: 224 interpolated points
Trying HE02GBR01_d18O:
Original data: 84 points, range 1568.0 to 1983.0
After cleaning: 82 points
Current median step: 5.000, target: 1.0
Interpolation range: 1568.0 to 1983.0
SUCCESS: 416 interpolated points
Trying HE02GBR01_SrCa:
Original data: 84 points, range 1568.0 to 1983.0
After cleaning: 82 points
Current median step: 5.000, target: 1.0
Interpolation range: 1568.0 to 1983.0
SUCCESS: 416 interpolated points
=== RESULTS ===
Successfully interpolated: 27 records
Failed: 0 records
We then combine the dataframe to include both the interpolated records as well as the originals. The total number should be original total + interpolated.
new_rows_dfs = [pd.DataFrame([row]) for row in new_rows]
df_with_interp = pd.concat([df_cleaned_flip] + new_rows_dfs, ignore_index=True)
print(f"✅ Corrected dataframe shape: {df_with_interp.shape}")
print(f" Original records: {len(df_cleaned_flip)}")
print(f" Interpolated records: {len(new_rows)}")
print(f" Total: {len(df_with_interp)} (should be {len(df_cleaned_flip) + len(new_rows)})")
# Verify we have the right number of interpolated records
interp_count = sum(df_with_interp['TSID'].str.contains('_interp', na=False))
print(f" Interpolated records in dataframe: {interp_count}")
✅ Corrected dataframe shape: (711, 22)
Original records: 684
Interpolated records: 27
Total: 711 (should be 711)
Interpolated records in dataframe: 27
Now we run the same process as we did in Option 1 to convert our new combined dataFrame into a proxyDatabase. We will once again save this resulting proxyDataBase as a pickle and a netCDF.
print(f"\n" + "="*50)
print("Converting to CFR ProxyDatabase...")
try:
interp_pdb = create_pdb(df_with_interp)
print(f"SUCCESS: ProxyDatabase created with {interp_pdb.nrec} records")
print("Proxy types:", interp_pdb.type_dict)
# Verify seasonality is preserved
print(f"\nSeasonality verification:")
seasonality_preserved = 0
for pid, record in interp_pdb.records.items():
if record.seasonality is not None:
seasonality_preserved += 1
print(f"Records with seasonality: {seasonality_preserved}/{interp_pdb.nrec}")
# Show a sample record
sample_pid = list(interp_pdb.records.keys())[0]
sample_record = interp_pdb.records[sample_pid]
print(f"\nSample CFR ProxyRecord:")
print(f" PID: {sample_record.pid}")
print(f" Type: {sample_record.ptype}")
print(f" Seasonality: {sample_record.seasonality}")
print(f" Time points: {len(sample_record.time)}")
print(f" Location: ({sample_record.lat:.2f}, {sample_record.lon:.2f})")
except Exception as e:
print(f"ERROR creating CFR ProxyDatabase: {e}")
import traceback
traceback.print_exc()
==================================================
Converting to CFR ProxyDatabase...
Creating CFR ProxyDatabase from enhanced DataFrame...
Processing 0/711...
Processing 100/711...
Processing 200/711...
Processing 300/711...
Processing 400/711...
Processing 500/711...
Processing 600/711...
Processing 700/711...
Created CFR ProxyDatabase with 711 records
SUCCESS: ProxyDatabase created with 711 records
Proxy types: {'ice.d18O': 51, 'tree.MXD': 59, 'tree.TRW': 355, 'lake.other': 45, 'marine.other': 60, 'documents': 15, 'lake.reflectance': 1, 'coral.SrCa': 36, 'coral.d18O': 64, 'ice.melt': 2, 'borehole': 3, 'speleothem.d18O': 7, 'coral.calc': 2, 'sclerosponge.d18O': 10, 'hybrid': 1}
Seasonality verification:
Records with seasonality: 711/711
Sample CFR ProxyRecord:
PID: Ant_030
Type: ice.d18O
Seasonality: [1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12]
Time points: 1220
Location: (-79.46, 247.91)
filtered_pdb_2 = interp_pdb.filter(by='dt', keys=(0,1.1))
len(filtered_pdb_2.records)
586
filtered_pdb_2.to_nc('./prev_data/pdb_w_seasonality_annual_interp.nc')
>>> Warning: this is an experimental feature.
>>> Warning: this is an experimental feature.
100%|██████████| 586/586 [00:21<00:00, 26.66it/s]
>>> Data before 1 CE is dropped for records: ['Arc_032', 'SAm_025', 'Arc_025', 'Arc_002', 'Aus_041', 'Arc_031', 'Eur_017', 'Arc_021', 'NAm_880', 'Ant_031dD', 'Ant_031', 'NAm_664', 'Ant_022', 'Arc_020', 'Eur_014', 'Arc_004', 'Eur_001_interp', 'NAm_3870_interp', 'Eur_008_interp', 'Ant_035_interp'].
ProxyDatabase saved to: ./prev_data/pdb_w_seasonality_annual_interp.nc
pickle_path = './prev_data/pdb_w_seasonality_annual_interp.pkl'
with open(pickle_path, 'wb') as f:
pickle.dump(filtered_pdb_2, f)
print("Saving with seasonality preservation using pickle...")
print(f"✓ Saved to pickle: {pickle_path}")
# Test loading from pickle
with open(pickle_path, 'rb') as f:
test_db_pickle = pickle.load(f)
print(f"✓ Loaded from pickle: {test_db_pickle.nrec} records")
# Verify seasonality is preserved in pickle
seasonality_pickle = {}
for pid, record in test_db_pickle.records.items():
if record.seasonality is None:
key = "None"
elif record.seasonality == list(range(1, 13)):
key = "Annual [1-12]"
elif record.seasonality == [6, 7, 8]:
key = "Summer [6, 7, 8]"
elif record.seasonality == [12, 1, 2]:
key = "Winter [12, 1, 2]"
else:
key = str(record.seasonality)
seasonality_pickle[key] = seasonality_pickle.get(key, 0) + 1
print(f"\nSeasonality distribution in pickle-loaded database:")
for season, count in sorted(seasonality_pickle.items(), key=lambda x: x[1], reverse=True)[:10]:
print(f" {season}: {count}")
# Show examples
print(f"\nExample records from pickle with their seasonality:")
count = 0
for pid, record in test_db_pickle.records.items():
if count >= 10:
break
if record.seasonality != list(range(1, 13)) and record.seasonality is not None:
print(f" {record.pid}: {record.ptype} -> {record.seasonality}")
count += 1
# Verify counts
pickle_custom_seasonality = sum(1 for r in test_db_pickle.records.values()
if r.seasonality is not None and r.seasonality != list(range(1, 13)))
pickle_none_count = sum(1 for r in test_db_pickle.records.values() if r.seasonality is None)
# Debugging checks
print(f"\nPickle verification:")
print(f" Records with custom seasonality: {pickle_custom_seasonality}")
print(f" Records with None seasonality: {pickle_none_count}")
print(f" Records with annual seasonality: {test_db_pickle.nrec - pickle_custom_seasonality - pickle_none_count}")
if pickle_custom_seasonality > 0:
print(f"\n✅ SUCCESS: Seasonality preserved in pickle format!")
print(f" Use pickle format for now until CFR NetCDF seasonality support is fixed.")
else:
print(f"\n⚠️ Still having issues with seasonality preservation")
print(f"\n" + "="*60)
print(f"File: {pickle_path}")
Saving with seasonality preservation using pickle...
✓ Saved to pickle: ./prev_data/pdb_w_seasonality_annual_interp.pkl
✓ Loaded from pickle: 586 records
Seasonality distribution in pickle-loaded database:
Summer [6, 7, 8]: 309
Annual [1-12]: 226
[6, 7]: 6
Winter [12, 1, 2]: 6
[8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7]: 5
[5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4]: 4
[9, 10, 11, 12, 1, 2, 3, 4]: 4
[7, 8, 9]: 3
[3, 4, 5]: 2
[7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6]: 2
Example records from pickle with their seasonality:
NAm_568: tree.MXD -> [6, 7, 8]
Arc_060: tree.MXD -> [6, 7, 8]
NAm_1876: tree.TRW -> [6, 7, 8]
NAm_1108: tree.TRW -> [6, 7, 8]
NAm_1120: tree.MXD -> [6, 7, 8]
Asia_102: tree.TRW -> [6, 7, 8]
SAm_025: lake.other -> [11, 12, 1, 2]
NAm_1396: tree.TRW -> [6, 7, 8]
NAm_220: tree.TRW -> [6, 7, 8]
SAm_020: tree.TRW -> [5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4]
Pickle verification:
Records with custom seasonality: 360
Records with None seasonality: 0
Records with annual seasonality: 226
✅ SUCCESS: Seasonality preserved in pickle format!
Use pickle format for now until CFR NetCDF seasonality support is fixed.
============================================================
File: ./prev_data/pdb_w_seasonality_annual_interp.pkl