Proxy Database Comparisons#
Comparing Interpolated vs. Non-Interpolated PReSto2k databases#
This comparison is straightforward. Since The only difference here should be the 27 records that got added in during the interpolation step, we can easily use cfr’s diff operator to directly subtract one from the other to identify the additional records. By doing this, if one reconstruction is significantly better or worse, we can more easily pinpoint which records are the cause.
print("Loading from pickle...")
with open('./prev_data/pdb_w_seasonality_annual.pkl', 'rb') as f:
no_interp = pickle.load(f)
print(f"Loaded {no_interp.nrec} records from pickle")
print("Loading from pickle...")
with open('./prev_data/pdb_w_seasonality_annual_interp.pkl', 'rb') as f:
with_interp = pickle.load(f)
print(f"Loaded {with_interp.nrec} records from pickle")
Loading from pickle...
Loaded 559 records from pickle
Loading from pickle...
Loaded 586 records from pickle
no_interp_pdb = no_interp
with_interp_pdb = with_interp
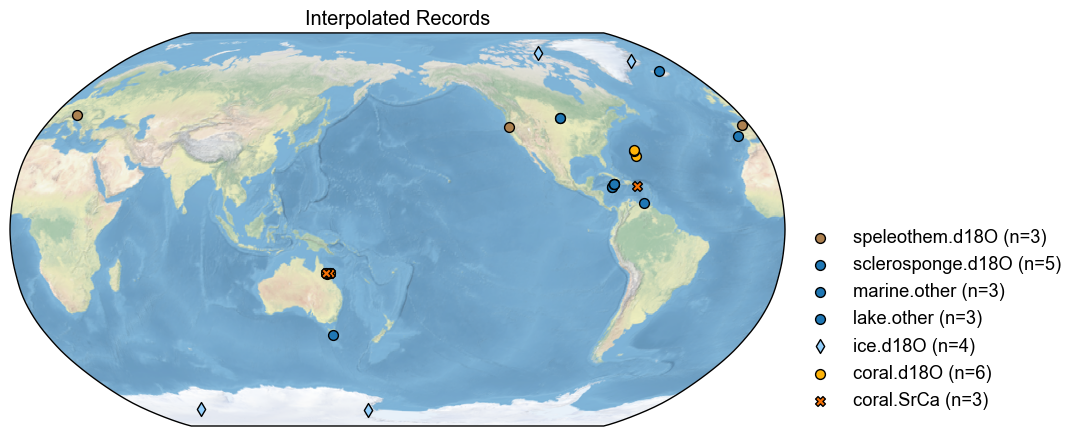
diff = with_interp_pdb-no_interp_pdb
diff.plot(title='Interpolated Records')
(<Figure size 1000x1000 with 1 Axes>,
{'map': <GeoAxes: title={'center': 'Interpolated Records'}>})
Comparing Original PAGES2k with PReSto2k#
This second comparison proves to be much more difficult. Here we compare the ProxyDatabase used in our reproduction of LMRv2.1.
Because the PID naming convention between PAGES2k in cfr and those pulled from the LiPDGraph are slightly different, despite possibly containing a majority of the same actual records, we have to create additional filters to catch all the similarities and differences. To bypass this without overcomplicating the names, we will use the following function match_pid_name to check each record between two proxy databases and make sure their lat, lon, and values match without regarding the PID.
# load original used in LMRv2.1 reproduction
pdb_og = cfr.ProxyDatabase().load_nc('./prev_data/pdb/filtered_ptype_res_cfr_pages2kv2.nc')
pdb_og.refresh()
We want the preferred pid map to be the pdb1 in the function, so when we use rename_pids, the no_interp_pdb names will change to the ones in pdb_og, which match the orignal naming convention used by cfr.
rename_pids matches proxy records between two proxy databases using a multi-pass strategy that becomes progressively more flexible. It starts with manual overrides for known pairs, then tries exact location + proxy type + value matches, followed by value-only matches, location-only matches with value prioritization, and finally pure value-based matches with stricter overlap requirements. The goal is to identify corresponding records across databases that may have different naming conventions but represent the same paleoclimate data, through as many steps as can filter out incorrect matches or duplicate matches.
import re
def _pid_num(pid):
"""
Extract the trailing numeric portion of a proxy record ID.
Parameters
----------
pid : str or int
Proxy record identifier (e.g., "Ocn_112"). Can be any object
convertible to string.
Returns
-------
int or None
The integer found at the end of the string if present,
otherwise None.
"""
m = re.search(r'(\d+)$', str(pid))
return int(m.group(1)) if m else None
def values_match(v1, v2, t1, t2, min_overlap_fraction=0.7, min_correlation=0.8):
"""
Test whether two proxy value series represent the same underlying record
based on overlapping time coverage and statistical similarity.
The function compares two time/value series over their common overlap
period. It checks (1) that enough of each record overlaps, and (2) that
the values are sufficiently correlated or numerically close.
Parameters
----------
v1, v2 : array_like
Value arrays for the two records.
t1, t2 : array_like
Time arrays corresponding to ``v1`` and ``v2``.
min_overlap_fraction : float, optional
Minimum fraction of overlap (by number of points) required between
the two series. Default is 0.7.
min_correlation : float, optional
Minimum Pearson correlation coefficient required when comparing
overlapping segments. Default is 0.8.
Returns
-------
bool
True if the two series are judged to match, False otherwise.
"""
if len(v1) == 0 or len(v2) == 0:
return False
# Find overlap
common_start = max(t1[0], t2[0])
common_end = min(t1[-1], t2[-1])
mask1 = (t1 >= common_start) & (t1 <= common_end)
mask2 = (t2 >= common_start) & (t2 <= common_end)
v1_overlap = v1[mask1]
v2_overlap = v2[mask2]
# More lenient overlap check
overlap_frac = min(len(v1_overlap) / len(v1), len(v2_overlap) / len(v2))
if overlap_frac >= min_overlap_fraction and len(v1_overlap) > 10:
if len(v1_overlap) == len(v2_overlap):
# Same length - use both correlation and closeness
if len(v1_overlap) > 1: # Need at least 2 points for correlation
corr = np.corrcoef(v1_overlap, v2_overlap)[0,1]
close = np.allclose(v1_overlap, v2_overlap, atol=1e-6, equal_nan=True)
return (not np.isnan(corr) and corr >= min_correlation) or close
else:
return np.allclose(v1_overlap, v2_overlap, atol=1e-6, equal_nan=True)
else:
# Different lengths - just check if they're close after interpolation to shorter length
min_len = min(len(v1_overlap), len(v2_overlap))
if min_len > 10:
# Simple downsampling to same length
v1_sampled = v1_overlap[::len(v1_overlap)//min_len][:min_len]
v2_sampled = v2_overlap[::len(v2_overlap)//min_len][:min_len]
if len(v1_sampled) == len(v2_sampled) and len(v1_sampled) > 1:
corr = np.corrcoef(v1_sampled, v2_sampled)[0,1]
return not np.isnan(corr) and corr >= min_correlation
return False
def match_proxy_records(pdb1, pdb2, tol_deg=0.03):
"""
Match proxy records between two databases using a multi-pass strategy.
Matching proceeds in several stages:
1. Apply manual overrides for known ID pairs.
2. Location + proxy type + value similarity, with PID numeric tiebreak.
3. Value-only matches (when no location match exists).
4. Location-only matches, prioritizing value similarity.
5. Pure value-based matches with stricter overlap requirements.
Parameters
----------
pdb1 : ProxyDatabase
First proxy database object. Must have a ``records`` dict mapping
record IDs to objects with attributes ``lat``, ``lon``, ``ptype``,
``time``, and ``value``.
pdb2 : ProxyDatabase
Second proxy database object, same requirements as ``pdb1``.
tol_deg : float, optional
Tolerance in degrees for considering two locations to be the same.
Default is 0.03 (~3 km).
Returns
-------
matches : dict
Mapping from record IDs in ``pdb1`` to their matched record IDs in ``pdb2``.
conflicts : dict
Dictionary of unresolved or conflicting matches. Keys are ``pdb2`` IDs,
values are lists of candidate ``pdb1`` IDs.
missing : list
List of record IDs in ``pdb1`` that could not be matched.
"""
matches, conflicts, missing = {}, {}, []
used_right = set()
# Manual overrides first
manual_matches = {
'Ocn_112': 'Ocean2kHR_100',
'Ocn_095': 'Ocean2kHR_156',
'Ant_019': 'Ant_033',
'Ocn_091': 'Ocean2kHR_157',
'Ocn_093': 'Ocean2kHR_158'
}
for pages_pid, presto_pid in manual_matches.items():
if pages_pid in pdb1.records and presto_pid in pdb2.records:
matches[pages_pid] = presto_pid
used_right.add(presto_pid)
# Pass 1: location + ptype + values + PID tiebreaker
for pid1, r1 in pdb1.records.items():
if pid1 in matches:
continue
cands = [pid2 for pid2, r2 in pdb2.records.items()
if (r2.ptype == r1.ptype and
abs(r1.lat - r2.lat) <= tol_deg and
abs(r1.lon - r2.lon) <= tol_deg and
pid2 not in used_right)]
if len(cands) == 1:
matches[pid1] = cands[0]
used_right.add(cands[0])
elif len(cands) > 1:
# Try values, then PID tiebreaker
value_cands = [pid2 for pid2 in cands
if values_match(
np.asarray(r1.value), np.asarray(pdb2.records[pid2].value),
np.asarray(r1.time), np.asarray(pdb2.records[pid2].time))]
if len(value_cands) == 1:
matches[pid1] = value_cands[0]
used_right.add(value_cands[0])
elif len(value_cands) > 1:
# PID tiebreaker
n1 = _pid_num(pid1)
if n1:
pid_match = [pid2 for pid2 in value_cands if _pid_num(pid2) == n1]
if len(pid_match) == 1:
matches[pid1] = pid_match[0]
used_right.add(pid_match[0])
# Pass 2: values-only
for pid1, r1 in pdb1.records.items():
if pid1 in matches:
continue
value_matches = [pid2 for pid2, r2 in pdb2.records.items()
if (pid2 not in used_right and
values_match(
np.asarray(r1.value), np.asarray(r2.value),
np.asarray(r1.time), np.asarray(r2.time)))]
if len(value_matches) == 1:
matches[pid1] = value_matches[0]
used_right.add(value_matches[0])
# Mark remaining as missing
for pid1 in pdb1.records:
if pid1 not in matches:
missing.append(pid1)
# Pass 3: Location-only with value priority
for pid1, r1 in pdb1.records.items():
if pid1 in matches:
continue
# Find all records at same location regardless of ptype
location_matches = []
for pid2, r2 in pdb2.records.items():
if (pid2 not in used_right and
abs(r1.lat - r2.lat) <= tol_deg and
abs(r1.lon - r2.lon) <= tol_deg):
location_matches.append(pid2)
if len(location_matches) == 1:
matches[pid1] = location_matches[0]
used_right.add(location_matches[0])
elif len(location_matches) > 1:
# Prioritize value matching
value_matches = [pid2 for pid2 in location_matches
if values_match(
np.asarray(r1.value), np.asarray(pdb2.records[pid2].value),
np.asarray(r1.time), np.asarray(pdb2.records[pid2].time))]
if len(value_matches) == 1:
matches[pid1] = value_matches[0]
used_right.add(value_matches[0])
elif len(value_matches) == 0:
# No value matches - leave unmatched rather than guess
continue
else:
# Multiple value matches - only use PID as very last resort
n1 = _pid_num(pid1)
if n1:
pid_matches = [pid2 for pid2 in value_matches if _pid_num(pid2) == n1]
if len(pid_matches) == 1:
matches[pid1] = pid_matches[0]
used_right.add(pid_matches[0])
# If still multiple candidates, leave unmatched rather than guess
# Pass 4: Pure value matching with substantial overlap
for pid1, r1 in pdb1.records.items():
if pid1 in matches:
continue
# Check all remaining records for value similarity
for pid2, r2 in pdb2.records.items():
if (pid2 not in used_right and
values_match(
np.asarray(r1.value), np.asarray(r2.value),
np.asarray(r1.time), np.asarray(r2.time),
min_overlap_fraction=0.8)): # Slightly more lenient
matches[pid1] = pid2
used_right.add(pid2)
break # Take first substantial match found
# Update missing list
missing = [pid for pid in pdb1.records if pid not in matches]
return matches, conflicts, missing
def rename_pids(pdb, matches, in_place=False):
"""
Rename record identifiers (PIDs) in a ProxyDatabase.
This function replaces PIDs in a ProxyDatabase according to a mapping of
`{new_pid: old_pid}`. Each record with an `old_pid` key is reassigned to
`new_pid`. If `new_pid` already exists in the database, that rename is skipped
to avoid collisions. Records not listed in the mapping remain unchanged.
Parameters
----------
pdb : cfr.ProxyDatabase
The ProxyDatabase whose records should be renamed.
matches : dict
Dictionary mapping `{new_pid: old_pid}`. Keys are the preferred PIDs to
assign, values are the current PIDs in `pdb`.
in_place : bool, optional
If True, rename records directly in `pdb`. If False (default), return a
modified copy and leave the original unchanged.
Returns
-------
cfr.ProxyDatabase
A ProxyDatabase with updated PIDs following the mapping. If `in_place`
is False, this is a new object; otherwise the input object is modified
and returned.
"""
db = pdb if in_place else pdb.copy()
recs = db.records
for new_pid, old_pid in matches.items():
if old_pid in recs and new_pid not in recs:
rec = recs.pop(old_pid)
# update the record's internal pid if it has one
if hasattr(rec, "pid"):
rec.pid = new_pid
recs[new_pid] = rec
return db
# Now test the matching
matches, conflicts, missing = match_proxy_records(pdb_og, no_interp_pdb)
print("=== MATCHING RESULTS ===")
print(f"Matches found: {len(matches)}")
print(f"Conflicts: {len(conflicts)}")
print(f"Missing from PReSto2k: {len(missing)}")
=== MATCHING RESULTS ===
Matches found: 505
Conflicts: 0
Missing from PReSto2k: 53
# Re-run the renaming with all matches
no_interp_fully_renamed = rename_pids(no_interp_pdb, matches, in_place=False)
Now that we have renamed the matching pids, we can use the Proxy Database subtraction that is built into cfr. This will give us two maps of which records are missing from both the PReSto2k database and the original PAGES2k database used for LMRv2.1.
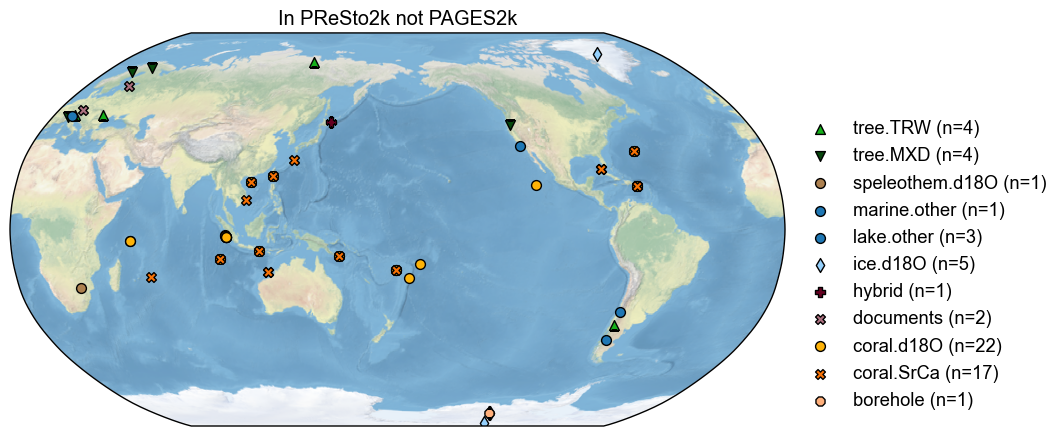
# Plot the map of records not in PAGES2k
diff = no_interp_fully_renamed - pdb_og
diff.plot(title='In PReSto2k not PAGES2k')
>>> Subtracting Ocn_167 failed.
>>> Subtracting Ant_024 failed.
>>> Subtracting Ocn_086 failed.
>>> Subtracting Ocn_076 failed.
>>> Subtracting Ocn_099 failed.
>>> Subtracting Ocn_158 failed.
>>> Subtracting Arc_035 failed.
>>> Subtracting Ocn_177 failed.
>>> Subtracting Ocn_171 failed.
>>> Subtracting Ocn_069 failed.
>>> Subtracting Ocn_170 failed.
>>> Subtracting NAm_088 failed.
>>> Subtracting Ocn_116 failed.
>>> Subtracting Ocn_130 failed.
>>> Subtracting Ocn_131 failed.
>>> Subtracting Ocn_147 failed.
>>> Subtracting Ocn_174 failed.
>>> Subtracting Ocn_160 failed.
>>> Subtracting Ocn_172 failed.
>>> Subtracting Arc_066 failed.
>>> Subtracting Ocn_166 failed.
>>> Subtracting Ocn_140 failed.
>>> Subtracting Ocn_141 failed.
>>> Subtracting Ocn_179 failed.
>>> Subtracting Ant_007 failed.
>>> Subtracting Ant_028 failed.
>>> Subtracting Eur_005 failed.
>>> Subtracting Ocn_168 failed.
>>> Subtracting Ocn_142 failed.
>>> Subtracting Ocn_143 failed.
>>> Subtracting Ocn_129 failed.
>>> Subtracting Ocn_120 failed.
>>> Subtracting Ocn_121 failed.
>>> Subtracting Ocn_127 failed.
>>> Subtracting Ant_008 failed.
>>> Subtracting Ocn_128 failed.
>>> Subtracting Ocn_173 failed.
>>> Subtracting Ocn_118 failed.
>>> Subtracting Ocn_139 failed.
>>> Subtracting Ocn_061 failed.
>>> Subtracting Ocn_062 failed.
>>> Subtracting Ocn_079 failed.
>>> Subtracting NAm_131 failed.
>>> Subtracting Ocn_107 failed.
>>> Subtracting Ocn_157 failed.
>>> Subtracting Ocn_146 failed.
>>> Subtracting Ocn_181 failed.
>>> Subtracting Ocn_182 failed.
>>> Subtracting Ocn_162 failed.
>>> Subtracting Ocn_163 failed.
>>> Subtracting Ocn_161 failed.
>>> Subtracting Ocn_125 failed.
>>> Subtracting Ocn_090 failed.
>>> Subtracting Ocn_119 failed.
>>> Subtracting Ocn_097 failed.
>>> Subtracting Ocn_159 failed.
>>> Subtracting Arc_024 failed.
>>> Subtracting Ocn_169 failed.
>>> Subtracting Ocn_071 failed.
>>> Subtracting Ocn_072 failed.
(<Figure size 1000x1000 with 1 Axes>,
{'map': <GeoAxes: title={'center': 'In PReSto2k not PAGES2k'}>})
# Plot the map of records not in PReSto2k
diff_other = pdb_og - no_interp_fully_renamed
diff_other.plot(title='In PAGES2k not PReSto2k')
>>> Subtracting SAm_020 failed.
>>> Subtracting LPDb9285123 failed.
>>> Subtracting Eur_026 failed.
>>> Subtracting SAm_0243 failed.
>>> Subtracting Eur_022 failed.
>>> Subtracting Ant_037 failed.
>>> Subtracting NAm_3100 failed.
>>> Subtracting SAm_032 failed.
>>> Subtracting Eur_002 failed.
>>> Subtracting Arc_031 failed.
>>> Subtracting Eur_017 failed.
>>> Subtracting Eur_016 failed.
>>> Subtracting Arc_058 failed.
>>> Subtracting Ant_031dD failed.
>>> Subtracting Ant_031 failed.
>>> Subtracting Eur_018 failed.
>>> Subtracting O2kLR_183 failed.
>>> Subtracting Ant_050 failed.
>>> Subtracting Asia_235 failed.
>>> Subtracting Eur_014 failed.
>>> Subtracting Arc_023 failed.
>>> Subtracting Africa_001 failed.
>>> Subtracting Ocean2kHR_011_iso2k failed.
>>> Subtracting KA17RYU01_SrCa failed.
>>> Subtracting FL17DTO02_SrCa failed.
>>> Subtracting BO14HTI01_SrCa failed.
>>> Subtracting SA16CLA01_d18O failed.
>>> Subtracting AB20MEN03_d18O failed.
>>> Subtracting AB20MEN02_d18O failed.
>>> Subtracting AB20MEN06_d18O failed.
>>> Subtracting AB20MEN07_d18O failed.
>>> Subtracting ZI16ROD01_SrCa failed.
>>> Subtracting AB20MEN05_d18O failed.
>>> Subtracting HE13MIS01_d18O failed.
>>> Subtracting HE13MIS01_SrCa failed.
>>> Subtracting AB20MEN04_d18O failed.
>>> Subtracting RA20TAI01_SrCa failed.
>>> Subtracting RA20TAI01_d18O failed.
>>> Subtracting TA18TAS01_d18O failed.
>>> Subtracting LI04FIJ01_d18O failed.
>>> Subtracting LI04FIJ01_SrCa failed.
>>> Subtracting WU13TON01_d18O failed.
>>> Subtracting CH97BVB01_d18O failed.
>>> Subtracting HE18COC01_d18O failed.
>>> Subtracting HE18COC01_SrCa failed.
>>> Subtracting MU18NPI01_SrCa failed.
>>> Subtracting MU18NPI01_d18O failed.
>>> Subtracting ZI15IMP02_SrCa failed.
>>> Subtracting KI14PAR01_SrCa failed.
>>> Subtracting KI14PAR01_d18O failed.
>>> Subtracting ZI15IMP01_SrCa failed.
>>> Subtracting GO08BER01_SrCa failed.
>>> Subtracting GO08BER01_d18O failed.
>>> Subtracting KI08PAR01_SrCa failed.
>>> Subtracting AB20MEN09_d18O failed.
>>> Subtracting AB20MEN08_d18O failed.
>>> Subtracting XI17HAI01_SrCa failed.
>>> Subtracting XI17HAI01_d18O failed.
>>> Subtracting DE14DTO01_SrCa failed.
>>> Subtracting DE14DTO02_SrCa failed.
>>> Subtracting LI06FIJ01_d18O failed.
(<Figure size 1000x1000 with 1 Axes>,
{'map': <GeoAxes: title={'center': 'In PAGES2k not PReSto2k'}>})
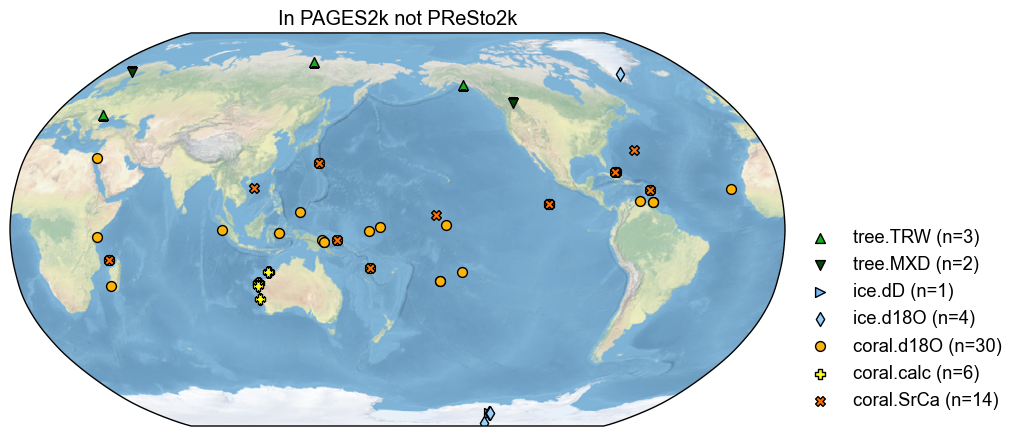
We see that the PReSto 2k criteria dropped a number of records, notably 6 coral calcification records from Western Australia. Multiple coral sites also appear to have been common to both PAGES 2k and CoralHydro2k, and might simply have been swapped from PAGES 2k to PReSto 2k.
print(f'There are {len(diff.records)} in PReSto2k but NOT in PAGES2k')
print(f'There are {len(diff_other.records)} in PAGES2k but NOT in PReSto2k')
There are 61 in PReSto2k but NOT in PAGES2k
There are 60 in PAGES2k but NOT in PReSto2k
Comparing Metadata vs. Class-Based Seasonality#
We will now run the same renaming/matching test on metadata-based and class-based seasonality. The two differ because not every record in these databases contains seasonality metadata. In this analysis, we chose to drop the records that do not contain this information. A more permissive approach would be to assign class-based seasonality to those records instead of dropping them altogether.
One important difference of note is that we are doing this comparison before calibration. The calibration step automatically filters out a number of records that do not meet certain requirements (See: Run Data Assimilation). Due to this, the class_og proxy database is unfiltered from the get-go (initial LiPD dataframe) and not filtered by resolution unlike the metadata ones (both with and without interpolation). During calibration, ~120 records get removed and hence this current proxy database appears significantly larger than what is actually assimilated in the DA step.
In a further experiment, it will be helpful to compare just the records that were used post-calibration between the two Proxy Databases to get a full breakdown for why one reconstruction outperforms the other.
class_og = cfr.ProxyDatabase().load_nc('./prev_data/lipd_pdb_pids.nc')
class_og.refresh()
matches_2, conflicts_2, missing_2 = match_proxy_records(class_og, no_interp_pdb)
# Re-run the renaming with all matches
no_interp_fully_renamed_2 = rename_pids(no_interp_pdb, matches_2, in_place=False)
diffo = class_og - no_interp_fully_renamed_2
diffo2 = no_interp_fully_renamed_2-class_og
>>> Subtracting Ocean2kHR_095 failed.
>>> Subtracting Ant_031dD failed.
>>> Subtracting Ocean2kHR_157 failed.
>>> Subtracting Ocean2kHR_158 failed.
>>> Subtracting FL17DTO02_SrCa failed.
>>> Subtracting AB20MEN03_d18O failed.
>>> Subtracting AB20MEN07_d18O failed.
>>> Subtracting LI04FIJ01_SrCa failed.
>>> Subtracting LI06FIJ01_d18O failed.
>>> Subtracting Ocn_066 failed.
>>> Subtracting Asi_324 failed.
>>> Subtracting Ocn_156 failed.
>>> Subtracting Ant_010 failed.
>>> Subtracting Arc_051 failed.
>>> Subtracting Ocn_045 failed.
>>> Subtracting Arc_040 failed.
>>> Subtracting Ocn-EmeraldBasin.Keigwin.2007_LPD68872399 failed.
>>> Subtracting Ant-DSS.DahlJensen.1999_LPD7abab671 failed.
>>> Subtracting NAm_3810 failed.
>>> Subtracting Ocn_194 failed.
>>> Subtracting Ocn_193 failed.
>>> Subtracting Ocn-MD98-2160.Newton.2010_LPD7e8e8eaa failed.
>>> Subtracting Ocn-WestSpitzberg.Bonnet.2010_LPD995c6f3c failed.
>>> Subtracting Ocn_074 failed.
>>> Subtracting NAm_3804 failed.
>>> Subtracting Ocn-FiskBasin.Richey.2009_LPD57d40973 failed.
>>> Subtracting Eur_024 failed.
>>> Subtracting Asi_230 failed.
>>> Subtracting Arc_044 failed.
>>> Subtracting Ocn-AlboranSea436B.Nieto-Moreno.2013_LPD0e0867fe failed.
>>> Subtracting Ocn_187 failed.
>>> Subtracting Asi_317 failed.
>>> Subtracting Eur_001 failed.
>>> Subtracting Asi_305 failed.
>>> Subtracting Ocn-CH07-98-MC-22.Saenger.2011_LPDa03ec713 failed.
>>> Subtracting NAm_3816 failed.
>>> Subtracting Ocn-FeniDrift.Richter.2009_LPD47182517 failed.
>>> Subtracting Ocn-WesternSvalbard.Spielhagen.2011_O2kLR_007 failed.
>>> Subtracting NAm_3851 failed.
>>> Subtracting Ant_009 failed.
>>> Subtracting Ocn-MD98-2177.Newton.2010_LPD30c5b95c failed.
>>> Subtracting Ocn-ODP984.Came.2007_LPDbd07f717 failed.
>>> Subtracting Asi_247 failed.
>>> Subtracting Eur_009 failed.
>>> Subtracting Asi_321 failed.
>>> Subtracting Ant_025 failed.
>>> Subtracting Ocn-GulfofGuinea.Weldeab.2007_LPD9cc3baf6-dup failed.
>>> Subtracting Ocn_030 failed.
>>> Subtracting Arc_073 failed.
>>> Subtracting Ocn-SouthChinaSea.Zhao.2006_LPD44b374e7 failed.
>>> Subtracting Ocn_082 failed.
>>> Subtracting Asi_328 failed.
>>> Subtracting Ocn-JacafFjord.Sepulveda.2009_LPD6315f935 failed.
>>> Subtracting Asi_320 failed.
>>> Subtracting Ocn_114 failed.
>>> Subtracting NAm_3801 failed.
>>> Subtracting Asi-SihailongwaLake.Chu.2011_LPDdc74a376 failed.
>>> Subtracting NAm_3870 failed.
>>> Subtracting Ant_013 failed.
>>> Subtracting Aus-DuckholeLake.Saunders.2013_LPD335d863f failed.
>>> Subtracting Arc_039 failed.
>>> Subtracting Ocn_150 failed.
>>> Subtracting Arc_042 failed.
>>> Subtracting Ocn-PigmyBasin.Richey.2015_LPD572f7cc3 failed.
>>> Subtracting Ocn-MD95-2011.Grimalt.2002_O2kLR_105 failed.
>>> Subtracting Ocn_125 failed.
>>> Subtracting Afr-LakeMalawi.Powers.2011_Malawi05 failed.
>>> Subtracting Ocn-MinorcaContourite.Moreno.2012_LPD10e9f8f7 failed.
>>> Subtracting Afr_004 failed.
>>> Subtracting NAm_3853 failed.
>>> Subtracting Asi_327 failed.
>>> Subtracting Ocn-CariacoBasin.Lea.2003_LPDcda1c277 failed.
>>> Subtracting Ocn_047 failed.
>>> Subtracting Asi_326 failed.
>>> Subtracting Afr-P178-15P.Tierney.2015_LPD53e179ad failed.
>>> Subtracting Afr-P178-15P.Tierney.2015_LPD81e53153 failed.
>>> Subtracting Ocn_101 failed.
>>> Subtracting Ocn-WesternAntarcticPeninsula.Shevenell.2011_LPD2a3997a7 failed.
>>> Subtracting NAm_3798 failed.
>>> Subtracting Asi_329 failed.
>>> Subtracting Ocn-TagusMudPatch.Abrantes.2005_LPDb7ba9166 failed.
>>> Subtracting Ant-WDC06A.Steig.2013_Ant_031dD failed.
>>> Subtracting Asi_319 failed.
>>> Subtracting NAm_3807 failed.
>>> Subtracting Ocn_165 failed.
>>> Subtracting Arc_056 failed.
>>> Subtracting Ocn_129 failed.
>>> Subtracting Ocn_128 failed.
>>> Subtracting Ocn-SouthAtlanticWestAfrica.Leduc.2010_LPDa77273bb failed.
>>> Subtracting Ocn-DryTortugas.Lund.2006_LPDf7bace53 failed.
>>> Subtracting Asi_325 failed.
>>> Subtracting Arc_036 failed.
>>> Subtracting Ocn-GreatBahamaBank.Lund.2006_LPD7c7e19c5 failed.
>>> Subtracting Eur_008 failed.
>>> Subtracting NAm_3867 failed.
>>> Subtracting NAm_3868 failed.
>>> Subtracting Ocn-WEqPacific.Stott.2007_O2kLR_131 failed.
>>> Subtracting Ocn-SWCoastOfIndia.Saraswat.2013_O2kLR_118 failed.
>>> Subtracting Arc_052 failed.
>>> Subtracting Ocn-EasternTropicalNorthAtlantic.Kuhnert.2011_LPD13a78f9b failed.
>>> Subtracting Ocn_126 failed.
>>> Subtracting Ocn_127 failed.
>>> Subtracting Ocn_175 failed.
>>> Subtracting Ocn_146 failed.
>>> Subtracting Ocn_195 failed.
>>> Subtracting Ocn-Philippines.Stott.2007_LPD1a0245b7 failed.
>>> Subtracting Ocn_148 failed.
>>> Subtracting Ant_012 failed.
>>> Subtracting Arc_037 failed.
>>> Subtracting Ocn-GreatBahamaBank.Richter.2006_LPDefb3d2b0 failed.
>>> Subtracting Arc_041 failed.
>>> Subtracting Arc_078 failed.
>>> Subtracting Ocn-RAPiD-12-1K.Thornalley.2009_LPDb17a5e9e-dup failed.
>>> Subtracting Arc_043 failed.
>>> Subtracting Eur_010 failed.
>>> Subtracting Arc_049 failed.
>>> Subtracting NAm_3819 failed.
>>> Subtracting Asi_335 failed.
>>> Subtracting Ocn-CariacoBasin.Black.2007_LPDe51d17e8 failed.
>>> Subtracting Ocn-AlboranSea384B.Nieto-Moreno.2013_LPDasan2012 failed.
>>> Subtracting Ant_035 failed.
>>> Subtracting Ocn_157 failed.
>>> Subtracting Ocn_158 failed.
>>> Subtracting Ocn-CapeGhir.McGregor.2007_LPD84ab67b9 failed.
>>> Subtracting Arc_079 failed.
>>> Subtracting Arc_053 failed.
>>> Subtracting Ocn_017 failed.
>>> Subtracting Arc_048 failed.
>>> Subtracting Arc_045 failed.
>>> Subtracting Ocn_135 failed.
>>> Subtracting Asi_318 failed.
>>> Subtracting CO00DRBE_Ocean2kHR_125_iso2k failed.
>>> Subtracting SS03HAJA_Ocean2kHR_128_iso2k failed.
>>> Subtracting FL17DTO02_FL17DTO02_SrCa failed.
>>> Subtracting AB20MEN03_AB20MEN03_d18O failed.
>>> Subtracting AB20MEN07_AB20MEN07_d18O failed.
>>> Subtracting DR00NBB01_DR00NBB01_d18O failed.
>>> Subtracting LI04FIJ01_LI04FIJ01_SrCa failed.
>>> Subtracting RO19PAR01_RO19PAR01_SrCa failed.
>>> Subtracting HE02GBR01_HE02GBR01_d18O failed.
>>> Subtracting HE02GBR01_HE02GBR01_SrCa failed.
>>> Subtracting KI08PAR01_KI08PAR01_SrCa failed.
>>> Subtracting LI06FIJ01_LI06FIJ01_d18O failed.
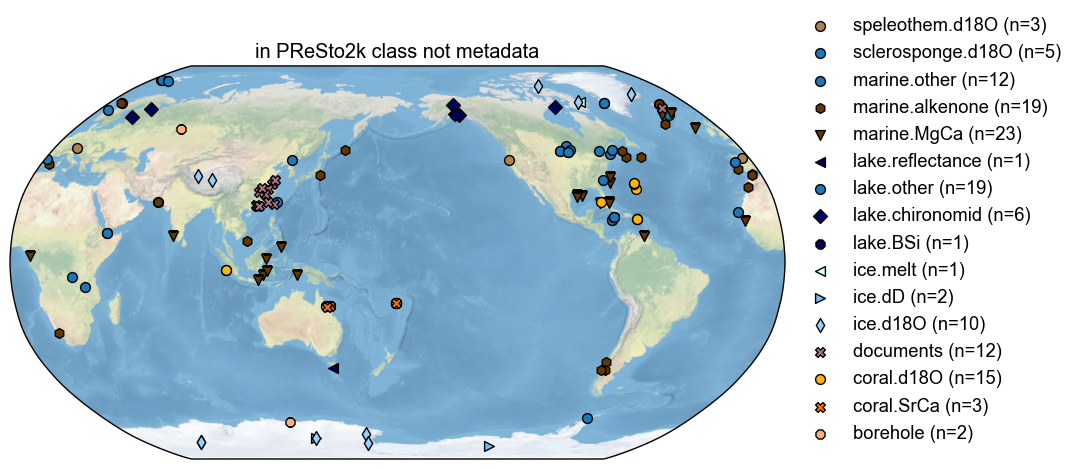
diffo.plot(title='in PReSto2k class not metadata')
(<Figure size 1000x1000 with 1 Axes>,
{'map': <GeoAxes: title={'center': 'in PReSto2k class not metadata'}>})
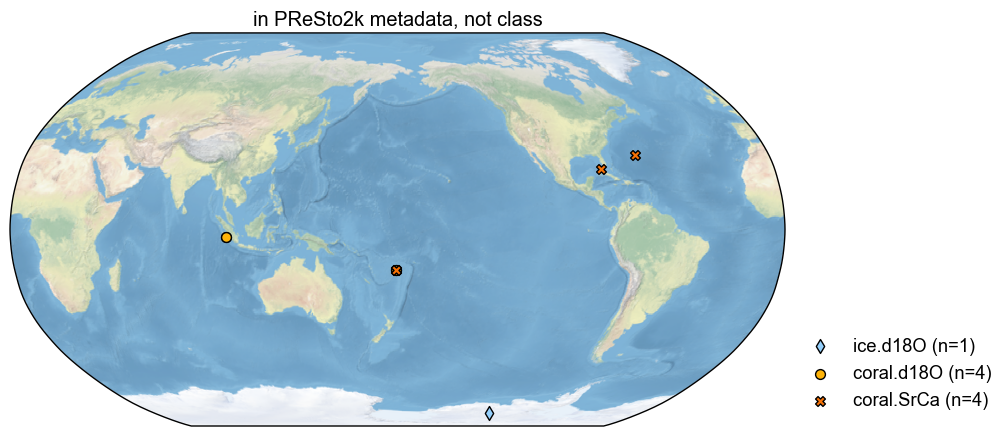
diffo2.plot(title='in PReSto2k metadata, not class')
(<Figure size 1000x1000 with 1 Axes>,
{'map': <GeoAxes: title={'center': 'in PReSto2k metadata, not class'}>})