Time Semantics#
Preamble#
Goals#
load timeseries data using Pandas
create a Pandas Series with desired time intervals
create Pyleoclim Series with desired time intervals
Plot Pyleoclim Series
Plot spectral analysis with Pyleoclim
Keywords#
Pre-requisites#
None. This tutorial assumes basic knowledge of Python. If you are not familiar with this coding language, check out this tutorial: http://linked.earth/LeapFROGS/.
Relevant Packages#
Pyleoclim, Pandas
Data Description#
Data provided by @bdamir5. “The data are my own measurements of CO2 concentration in the urban area of city Zagreb. They are sampled at a two-second frequency with the common low-cost CO2 sensor Senseair K30 and saved in .txt or .log files. Then I did some Python gymnastics to load all those log files to Pandas dataframe and extract whole-hour measurements. I exported the data to Excel to have them in one place when uploading here.”
Demonstration#
First import all necessary packages
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from matplotlib import dates as mdates
from matplotlib.ticker import FuncFormatter
import os
import pyleoclim as pyleo
import datetime
Load dataset#
Load dataset (excel file) as Pandas dataframe and print contents to see format.
df = pd.read_excel('../data/df_senzor2_cvetkovic.xlsx')
df.head()
| Time | CO2 Value [ppm] | |
|---|---|---|
| 0 | 2022-08-21 20:00:00 | 406 |
| 1 | 2022-08-21 21:00:00 | 412 |
| 2 | 2022-08-21 22:00:00 | 420 |
| 3 | 2022-08-21 23:00:00 | 439 |
| 4 | 2022-08-22 00:00:00 | 437 |
Convert Time column into datetime format (if not already)#
# convert time to datetime
df['Time'] = pd.to_datetime(df['Time'])
Create Pandas Series object from dataframe#
makes Series with datetime time as the index and CO2 ppm as the values. Then plot Pandas series.
#create series
data_series = pd.Series(data=df['CO2 Value [ppm]'].values,
index=df['Time'],
name = 'CO2 Value')
print(data_series)
Time
2022-08-21 20:00:00 406
2022-08-21 21:00:00 412
2022-08-21 22:00:00 420
2022-08-21 23:00:00 439
2022-08-22 00:00:00 437
...
2024-08-06 13:00:00 396
2024-08-06 14:00:00 391
2024-08-06 15:00:00 392
2024-08-06 16:00:00 393
2024-08-06 17:00:00 392
Name: CO2 Value, Length: 9677, dtype: int64
data_series.plot(title='CO2 ppm')
<Axes: title={'center': 'CO2 ppm'}, xlabel='Time'>
Create Pyleo Series with desired time frequency#
Create metadata with time unit and value unit to convert directly from Pandas Series to Pyleoclim Series. Then plot with Pyeloclim. You will notice that the time values change from YYYY-MM format into decimal year. This is done by default in pyleoclim to handle most paleoclimate data.
metadata = {
'time_name': 'Year',
'time_unit': 'year',
'value_name': data_series.name,
'value_unit': 'ppm'
}
# Convert to Pyleoclim Series
pyleo_series = pyleo.Series.from_pandas(
ser=data_series,
metadata=metadata
)
pyleo_series.plot(title='CO2 ppm')
(<Figure size 1000x400 with 1 Axes>,
<Axes: title={'center': 'CO2 ppm'}, xlabel='Time [year]', ylabel='CO2 Value [ppm]'>)
Changing plotting styles with Pyleoclim#
# try web for web-like font
pyleo.utils.plotting.set_style('web')
pyleo_series.plot(title='CO2 ppm')
(<Figure size 1000x400 with 1 Axes>,
<Axes: title={'center': 'CO2 ppm'}, xlabel='Time [year]', ylabel='CO2 Value [ppm]'>)
# back to the default -- matplotlib
pyleo.utils.plotting.set_style('matplotlib')
pyleo_series.plot(title='CO2 ppm')
(<Figure size 1000x400 with 1 Axes>,
<Axes: title={'center': 'CO2 ppm'}, xlabel='Time [year]', ylabel='CO2 Value [ppm]'>)

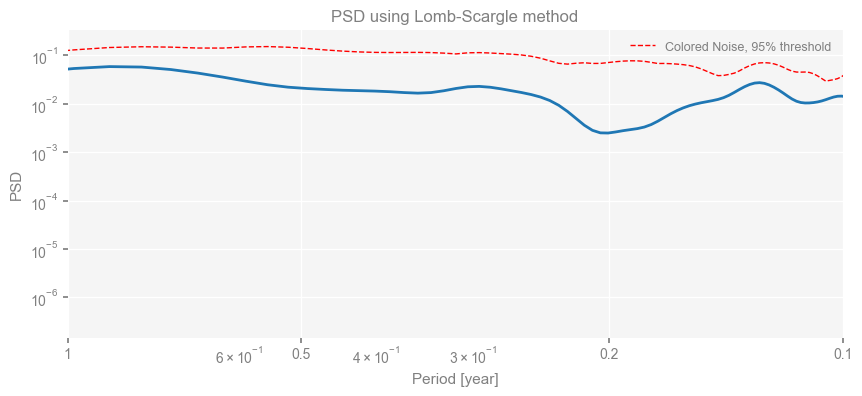
Spectral Analysis#
Using pyleoclim.Series.spectral() to plot the PSD and significance test
psd = pyleo_series.spectral(method='lomb_scargle')
psd_sig = psd.signif_test(method='CN',number=10) # This number should be much larger for actual science applications
Performing spectral analysis on individual series: 100%|██████████| 10/10 [01:02<00:00, 6.25s/it]
fig, ax = psd_sig.plot(title='PSD using Lomb-Scargle method')